Figures & data
Figure 1. General layout for transposable element (TE)-derived tandem repeat (TR) expansion and heterochromatin formation. (A) At some point in the TE life cycle one of its copies inserts into an active piRNA cluster and then serves as a template for the generation of complementary piRNAs that silence homologous sequences in the genome via heterochromatin formation. (B) The heterochromatinized TE copies harboring internal TRs are prone to suffering unequal recombination. This TR concertina generates variation in the size of heterochromatin blocks what may in turn affect gene expression. TIR: Terminal Inverted Repeats; LTR: Long Terminal Repeats.
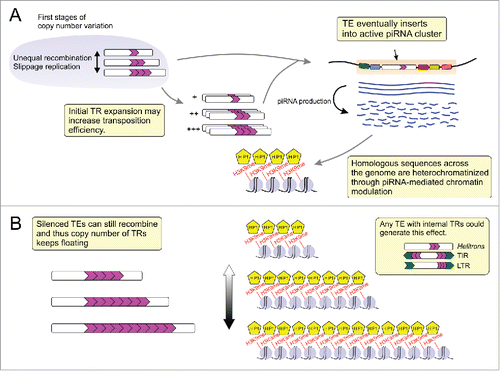
Figure 2. Schematic representation of the tandem insertions observed by Mendiola et al.Citation32 When the termination signal is missing at IS91 terminus, the RC transposition loops-out the entire plasmid and generate TIs of the whole construct (IS91 + pSU2572).
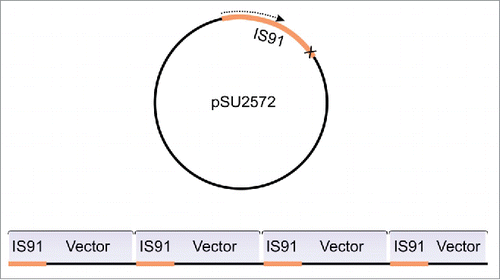
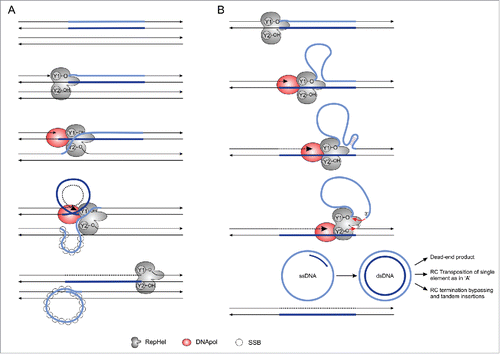
Figure 3. Two RC transposition models proposed for Helitrons. (A) The “Concerted” model of RC transposition was proposed for the IS91 prokaryotic elements and used to explain Helitron transposition.Citation5,39 Redrawn from Garcillán-Barcia et al.Citation41 (B) The “Sequential” model of RC transposition was proposed to explain the formation of episomal circular intermediates of IS91. Schematic representation based upon the model described in Garcillán-Barcia et al.Citation41 Single-stranded binding Proteins (SSBs) were not represented in the B section to improve clarity. RepHel: Replication Initiator Protein and Helicase; DNA Pol: host DNA polymerase.