Figures & data
Figure 1. Age-dependent increases in L1 in the mouse hippocampus genome. (A) Age-related variation in the copy numbers in the hippocampus (left panel) and frontal cortex (right panel). Six groups (3-, 8-, 15-, 18-, 20-, and 55-week-old) of C57BL/6J inbred mice were subjected to quantitative polymerase chain reaction (qPCR) analysis. The copy numbers of L1-ORF2 were measured using the 5S ribosomal gene as a reference. We analyzed a total of 42 mice (21 female, 21 male) at 3 (3 female, 3 male), 8 (6 female, 6 male), 15 (3 female, 3 male), 18 (3 female, 3 male), 20 (3 female, 3 male) and 55 (3 female, 3 male) weeks of age. The error bars show the standard error of the mean (SEM) values. The copy numbers in the hippocampus were significantly higher in 8-week-old inbred mice (Steel–Dwass test, 3-week-old vs 8-week-old, P = 0.008; 8-week-old vs 15-week-old, P = 0.4; 8-week-old vs 18-week-old, P = 0.08; 8-week-old vs 20-week-old, P = 0.0001). (B) The copy numbers of L1-5′UTR and -ORF1. The measurement analysis was done using the 5S ribosomal gene as a reference. A total of 18 mice (9 female, 9 male) were analyzed at 3 (3 female, 3 male), 8 (3 female, 3 male), and 20 (3 female, 3 male) weeks of age. Bars show the SEM. The copy numbers in the hippocampus were significantly higher in 8-week-old inbred mice (Tukey's test, 5′UTR; P = 0.0001, ORF1; P = 0.00003). (C) Stavudine (d4T)-induced inhibition of the age-dependent L1 increase in the hippocampus genome. Mice were administered d4T in drinking water beginning 3 weeks after birth and continued for 5 weeks, and then the L1 copy numbers in the hippocampus genome were analyzed. Bars show the SEM. The analysis included 47 control mice (3-week-old; 24 female, 8 male, 8-week-old; 9 female, 6 male) and 12 d4T-treated mice (3 female, 9 male). Of note, d4T treatment inhibited the L1 increase in 8-week-old mice (Steel–Dwass test, P = 0.0000009). (D) Representative results of the immunohistochemical analysis of d4T-treated mice. Ki67 (red; arrowheads), calretinin (green), 5-ethynyl-2-deoxyuridine (EdU) (red) and calretinin-EdU double-positive cells (yellow; arrowheads, inset) were visualised in the dentate gyrus of d4T-treated mice. Nuclei are depicted in blue. Scale bar, 50 µm. (E) Comparison of Ki67-positive cells and calretinin-EdU double-staining cells in mice with or without d4T treatment. Under graph shows calretinine-EdU double-staining cells (blue column) and EdU-positive cells (red column). The analysis included a total of 16 mice (8 female, 8 male); 8 control mice (4 female, 4 male) and 8 d4T-treated mice (4 female, 4 male). Ki67-positive cells and calretinin-EdU double-staining cells were counted in 3 randomly selected sections that were prepared from each mouse.
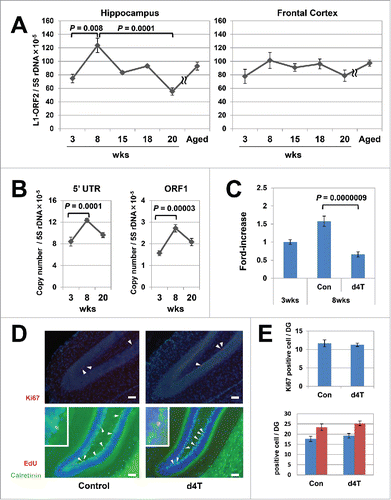
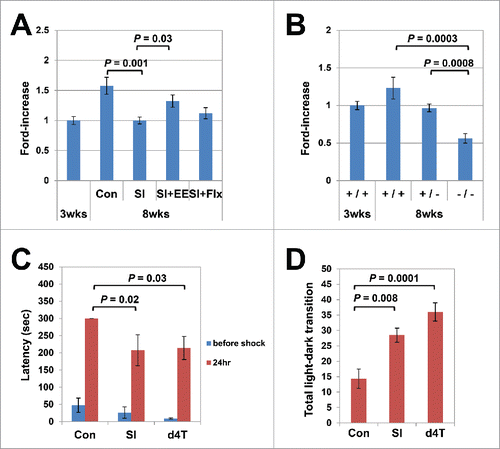
Figure 2. Age-dependent L1 increase in the hippocampus genome coupled with brain maturation. (A) Effects of social isolation (SI) and environmental enrichment (EE) on the age-dependent L1 increase. Mice were raised under SI stress for 5 weeks, beginning 3 weeks after birth, and the L1 copy numbers in the hippocampus genome were measured (Steel–Dwass test, group housing vs. SI; P = 0.001). The effects of fluoxetine treatment (Flx) and EE on the L1 copy numbers were analyzed under the same conditions. The analysis included a total of 35 mice (18 female, 17 male); 14 mice (8 female, 6 male) were raised under SI conditions, 11 (5 female, 6 male) under SI + EE conditions, and 10 (5 female, 5 male) under SI+Flx treatment. EE, but not Flx, restored the SI-induced inhibition of the age-dependent L1 increase (Steel–Dwass test, P = 0.03). The average running distance in the EE group was 23.28 km/day. (B) Inhibitory effects of mice lacking the NR2A. A total of 27 mice (13 female, 14 male): 13 wild-type mice (3-week-old; 3 female, 3 male, 8-week-old; 3 female, 4 male), 8 hetero-knockout (8-week-old; 4 female, 4 male) and 6 null mutant (8-week-old; 3 female, 3 male) mice with the same genotypes were raised in the same cages, and the L1 copy numbers in the hippocampus genome were measured at 8 weeks of age (Steel–Dwass test, wild-type vs. null knockout, P = 0.0003, wild-type vs. null hetero-type, P = 0.0008). (C) d4T administration induced abnormalities in passive avoidance. The response latency in the passive avoidance test was significantly shortened in the d4T-treated mice (Steel–Dwass test, P = 0.03). Again, the results were similar to those for the post-weaning SI mice (P = 0.02). (D) d4T administration induced abnormalities in the light/dark transition. The light/dark transition increased significantly in the d4T-treated mice (Tukey's test, P = 0.0001). The results were similar to those for the post-weaning SI mice (P = 0.008). The behavioral assay included a total of 21 mice (6 female, 15 male): 6 control mice (3 female, 3 male), 9 d4T-treated mice (3 female, 6 male), and 6 SI-treated mice (0 female, 6 male).