Figures & data
Figure 1. White gene TALEN design and mutagenesis. (A) The white gene locus on X chromosome and design of white TALEN pair 1 (w1 TALEN) and pair 2 (w2 TALEN). w1 TALEN targets the junction between exon 5 (yellow) and intron 5 (green). w2 TALEN targets the junction between intron 4 and exon 5. w2 TALEN arms (left and right) are represented as simplified RVD repeats: each colored oval representing a particular RVD recognizing a given nucleotide, the DNA recognition sequences are highlighted in matching color, and the conserved splicing acceptor dinucleotide 5′-AG-3′ sequence is highlighted in magenta. (B) Nature of mutations introduced by w2 TALEN. Among the 10 independent single G1 flies, 9 showed short deletion mutations of varying lengths (7-, 10-, and 11-nucleotide deletions). One G1 fly showed a short insertion of 16 nucleotides (pink).
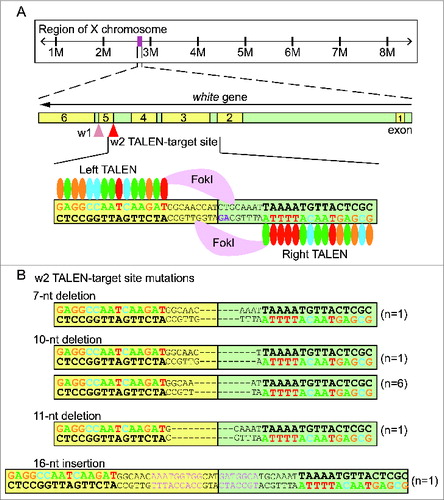
Figure 2. w2 TALEN-introduced somatic mutations and germline transmission. (A) The scheme of eye-color change introduced by w2 TALEN. From parental red-eyed flies (P), embryos injected w2 TALEN developed to G0 adults with mosaic eyes containing white patches. In G1 generation, white-eyed flies carried germline-transmitted mutations. (B) G0 somatic mutation rates at various w2 TALEN concentrations quantified by mosaic-eye phenotype. Numbers of G0 flies counted are listed on top of each bar. (C) The rational of quantifying global somatic mutations in individual G0 flies using SURVEYOR nuclease assay. Re-annealed amplicons around the predicted cut site undergone SURVEYOR nuclease treatment. The insert shows a representative result of digested product separation using Fragment Analyzer. Peaks at lower molecular weights (green) represent shorter DNA fragments generated by nuclease cut at a specific mutated site. See Figure S1 for more analysis and quantifications of gene modification rate. (D) G1 germline transmission rates at various w2 TALEN concentrations. The rate was represented by the percentage of white-eyed flies among total number of G1 progeny at each condition. Numbers of G1 flies counted are listed on top of each bar. (E) Comparisons of G1 germline transmission rates between progenies of non-mosaic eye (black bars) and mosaic eye G0 flies (green bars) at various w2 TALEN concentrations. Numbers of G1 flies counted are listed on top of each bar. (F) G0 larval survival rates at various w2 TALEN concentrations. This rate was represented by the percentage of injected embryos developed into first-instar larvae within 48 hours post injection from either wild-type (black bars) or lig4169 embryos (orange bars). Numbers of G0 embryos counted are listed on top of each bar.
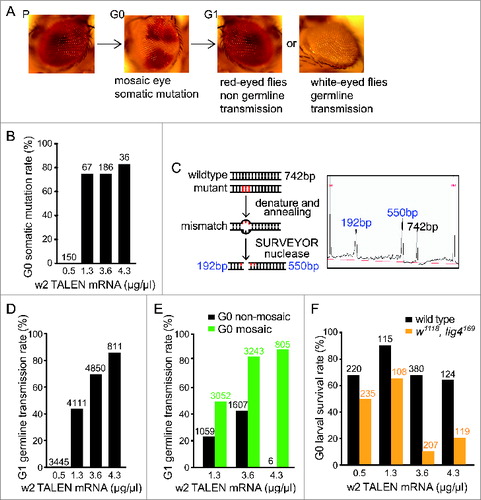
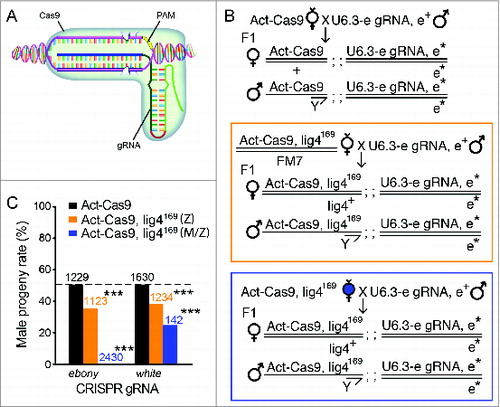
Figure 3. Cleavage-dependent lig4169 embryo lethality with the CRISPR/Cas9 system. (A) A cartoon diagram of the CRISPR/Cas9 system, which recognizes specific DNA using a RNA/DNA/protein complex (modified from our recent review articleCitation61). The 5′ end of gRNA sequence is used for target recognition on genomic DNA around a tri-nucleotide protospacer adjacent motif (PAM). Two tooth-shape structures represent Cas9 endonuclease active sites responsible for DNA cleavage on either stand of the double strand DNA. (B) Fly genetic crossing schemes for ubiquitous gene targeting using endogenously expressed Cas9 (Act-Cas9) and gRNAs (U6.3-ebony gRNA as an example here). F1 progenies bearing both transgenes activate the CRISPR/Cas9 system ubiquitously. Bi-allelic cuts on the target gene disrupt both copies of the locus (ebony, donated with asterisk). On the middle and bottom panel, lig4169 was recombined to Act-Cas9 chromosome to assay the contribution of Lig4 in animal survival in the gene-targeted progenies. While gene targeting is predicted to be equivalently efficient in progenies of either sex, the male progeny alone in both cases are zygotic hemizygous for lig4169 mutation. Sharing the same progeny genotypes, the middle (orange box) and bottom panel (blue box) differ by their maternal genotypes: heterozygous of lig4169 for the middle panel, and homozygous of lig4169 for the bottom panel. The homozygous mothers generate both maternal and zygotic null male progenies for Lig4. (C) The percentage of male progenies surviving ubiquitous CRISPR-mediated gene targeting 2 independent loci (ebony or white gRNAs), using Act-Cas9 (blank bars); Act-Cas9, lig4169 zygotic mutant only (Z, orange bars) or Act-Cas9, lig4169 maternal and zygotic mutant (M/Z, blue bars). The statistic deviation from the theoretical 50% percent perfect ratio was tested by student t-test (***P < 0.0001).