Figures & data
Figure 1. Cellular bioenergetics and mitochondrial function in 3T3-L1 fibroblasts and adipocytes. (A) Cellular bioenergetics measured by cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in 3T3-L1 fibroblasts and adipocytes. (B) Mitochondrial function represented by basal mitochondrial respiration (basal), respiration due to ATP turnover, uncoupled respiration (UCR) and maximal respiratory capacity in 3T3-L1 fibroblasts and adipocytes. (C) Uncoupled respiration as a percentage of basal mitochondrial respiration in 3T3-L1 fibroblasts and adipocytes. Data are represented as mean ± SEM, n = 10 biological replicates per group. † Denotes significantly different from fibroblasts for both OCR and ECAR. * Denotes significantly different from fibroblasts.
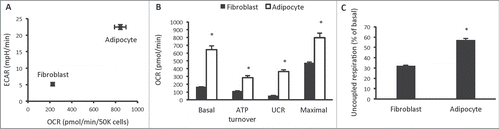
Figure 2. Gene expression profiling in 3T3-L1 adipocytes, white and brown adipose tissue. (A) Expression of genes that distinguish white adipocytes in 3T3-L1 adipocytes, epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT) and brown adipose tissue (BAT; subscapular brown fat). (B) Expression of genes that distinguish beige adipocytes in 3T3-L1 adipocytes, WAT and BAT. (C) Expression of genes that distinguish brown adipocytes in 3T3-L1 adipocytes, WAT and BAT. Data are represented as mean ± SEM, n = 6 biological replicates/individual mouse samples per group. * Denotes significantly different from all other groups.
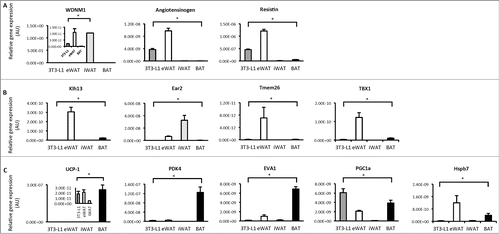
Figure 3. 3T3-L1 adipocytes increase oxygen consumption and the expression of brown adipocyte genes in response to norepinephrine. (A) Basal cellular oxygen consumption rate (OCR) in 3T3-L1 adipocytes treated with 100 μM GDP (UCP1 inhibitor) or 5 μM CsA (inhibitor of mitochondrial transition pore opening) for 24 hrs. (B) Phosphorylation of PKA T197, CREB S133, HSL S563 and total UCP-1 and tubulin in 3T3-L1 adipocytes exposed to vehicle (H2O) or norepinephrine (1 μM) for 20 min. (C) UCP1 gene expression in 3T3-L1 adipocytes following exposure to vehicle (H2O) or norepinephrine (1 μM) for 60 min. (D) OCR immediately following acute exposure to vehicle (H2O) or norepinephrine (1 μM) in 3T3-L1 adipocytes that had been previously treated with vehicle (0.1% DMSO), 100μM GDP or 5 μM CsA for 24 hrs. (E) Expression of genes that discriminate white, beige and brown adipocytes in 3T3-L1 adipocytes treated with norepinephrine (1 μM) for 60 min relative to control (vehicle) treated cells. Data are represented as mean ± SEM, n = 3 −6 biological replicates per group. * Denotes significantly different from vehicle treated cells.
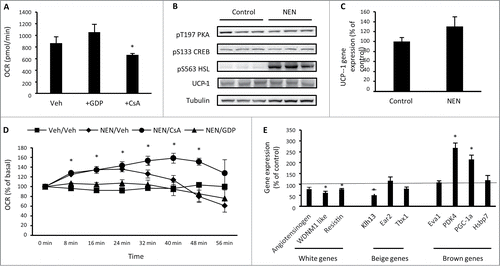
Figure 4. Chronic norepinephrine exposure in 3T3-L1 adipocytes. (A) Cellular bioenergetics measured by cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in 3T3-L1 adipocytes treated with vehicle (H2O; Control) or 1 μM norepinephrine for 48 hrs (ChNEN). (B) Mitochondrial function represented by basal mitochondrial respiration (basal), respiration due to ATP turnover, uncoupled respiration (UCR) and maximal respiratory capacity in Control and ChNEN 3T3-L1 L1 adipocytes. (C) Uncoupled respiration as a percentage of basal mitochondrial respiration in in Control and ChNEN 3T3-L1 adipocytes. (D) Basal cellular oxygen consumption rate (OCR) in Control and ChNEN 3T3-L1 adipocytes that were co-treated with vehicle (0.1% DMSO), 100 μM GDP or 5 μM CsA in the final 24 hrs. (E) Total UCP-1 and tubulin protein in Control and ChNEN 3T3-L1 L1 adipocytes. (F) OCR immediately following acute exposure to vehicle (H2O) or norepinephrine (1 μM) in Control and ChNEN 3T3-L1 adipocytes. (G) OCR immediately following acute exposure to vehicle (H2O) or norepinephrine (1 μM) in Control and ChNEN 3T3-L1 adipocytes that were co-treated with vehicle (0.1% DMSO), 100 μM GDP or 5 μM CsA in the final 24 hrs. Data are represented as mean ± SEM, n = 6 biological replicates per group. ∫ Denotes significantly different from Control cells for OCR. * Denotes significantly different from vehicle treated and control cells.
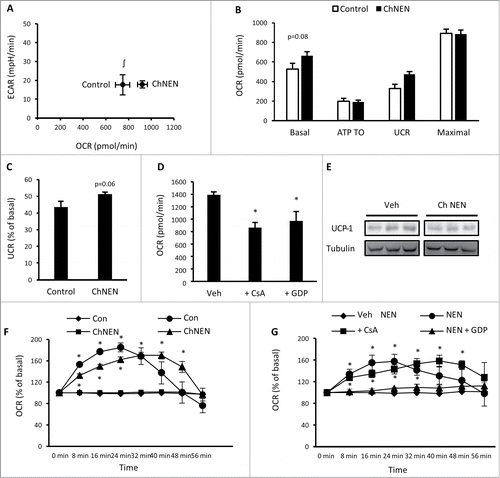
Figure 5. Bioenergetics and gene expression in 3T3-L1 adipocytes following differentiation without phosphodiesterase inhibitors. (A) Cellular bioenergetics measured by cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in 3T3-L1 cells following normal adipogenic differentiation (Norm. diff.), insulin only differentiation (Ins. diff.), or beige adipocyte differentiation (Beige diff.) (B) Mitochondrial function represented by basal mitochondrial respiration (basal), respiration due to ATP turnover, uncoupled respiration (UCR) and maximal respiratory capacity in Norm. diff., Ins. diff. and Beige diff. 3T3-L1 adipocytes. (C) Expression of genes that discriminate white, beige and brown adipocytes in Norm. diff. and Ins. diff. 3T3-L1 adipocytes. Data are represented as mean ± SEM, n = 6 −10 biological replicates per group. † Denotes significantly different from Ins. diff. cells for both OCR and ECAR. * Denotes significantly different from Norm. diff. # Denotes significantly different from Norm. diff and Ins. diff.
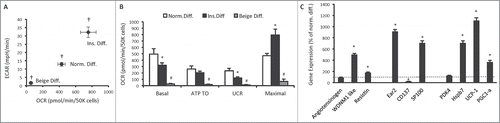
Table 1. Real time RT-PCR primer sequences
