Figures & data
Figure 1. Inhibition of Wnt signaling pharmacologically enhances browning (A) Experiment schematics of in vitro browning experiment. Primary mouse adipocytes differentiated from the stromal vascular fraction (SVF) of inguinal fat pads were treated with 10 nM of C59 for 6 d from the start of differentiation. Browning was induced by adding 1 µM norepinephrine. (B-D) qPCR analysis of Wnt signaling gene (Axin2) (B), BAT genes (Ucp1, Cidea, Ppargc1a) (C), and pan-adipocyte genes (Pparg, Fabp4) (D) on RNA extracted at Day 6 according to scheme 1(A). Shown is the relative mRNA expression values normalized with the housekeeping gene Rpl23. All data are presented as mean ± SEM by Student's t-test. * p < 0.05, ** p < 0.01 and *** p < 0.001 comparing with control. The fold of induction or inhibition is indicated when the change is significant and relevant. (E) Oil red O staining of primary mouse adipocytes at Day 6.
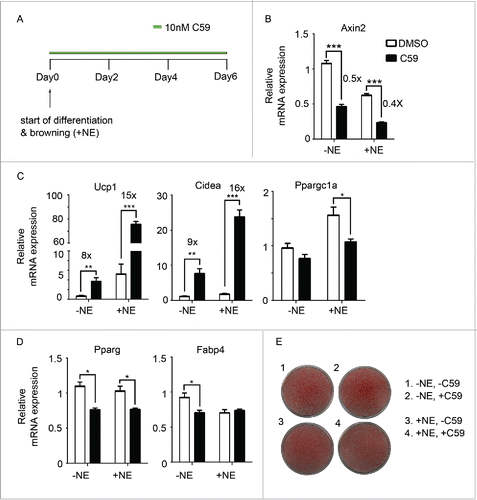
Figure 2. Inhibition of Wnt signaling genetically enhances browning (A) Porcn deletion in Rosa26-CreERT2/Porcnfl/fl mice upon addition of 4-hydroxytamoxifen (4-OHT). Δ: PCR product after Porcn is deleted. (B-D) SVF was isolated from inguinal fat pads of Rosa26-CreERT2/Porcnfl/fl mice and differentiated to adipocytes in the presence or absence of 4-OHT to delete Porcn. RNA was extracted at Day 6 of differentiation. qPCR analysis of Wnt signaling gene (Axin2) (B), BAT genes (Ucp1, Cidea, Ppargc1a) (C), and pan-adipocyte genes (Pparg, Fabp4) (D) on RNA extracted at Day 6 according to scheme 1(A). Shown is the relative mRNA expression values normalized with the housekeeping gene Rpl23. The fold of induction or % of inhibition is indicated when the change is significant and relevant. All data are presented as mean ± SEM by Student's t-test. * p < 0.05, ** p < 0.01 and *** p < 0.001 comparing with the control model.
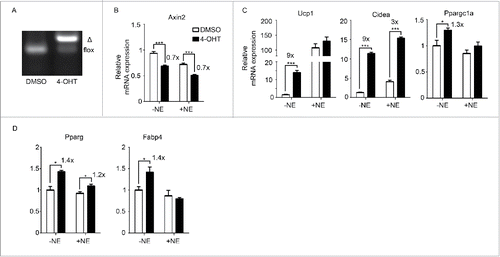
Figure 3. Wnt inhibition likely targets beige precursor cells to enhance browning (A) Primary adipocytes were treated with a second Wnt signaling inhibitor, XAV939. Shown is the qPCR analysis of marker genes after treatment following scheme 1(A). (B) Experiment schematics of in vitro browning experiment using primary adipocytes. Cells were treated with 100 nM XAV939 for only 2 d starting from Day 0, Day 2 or Day 4 during differentiation. RNA was harvested at Day 6 for qPCR analysis. (C) qPCR analysis of markers genes according to experiment plan 3 (B). (D) qPCR analysis of markers genes according to experiment plan 3(B) using 3T3-L1. Shown is the relative mRNA expression values normalized with housekeeping genes Rpl23. The fold of induction is indicated when the change is significant and relevant. All data are presented as mean ± SEM by Student's t-test. * p < 0.05, ** p < 0.01 and *** p < 0.001 comparing with control.
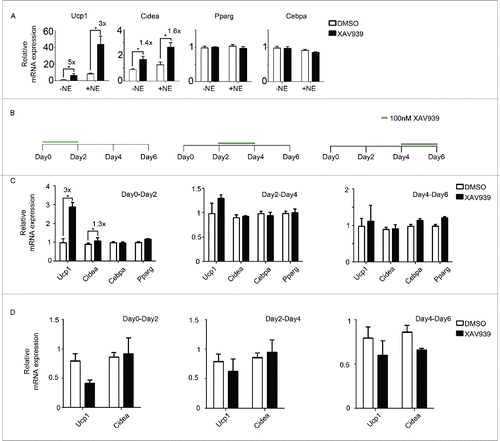
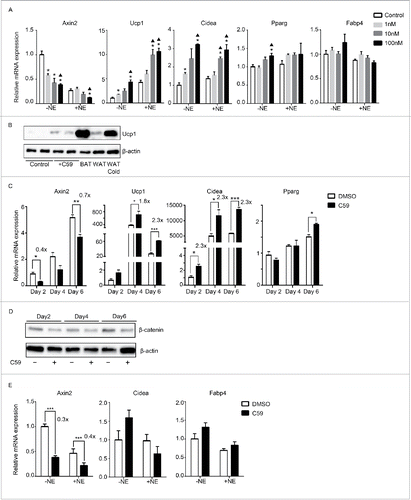
Figure 4. Wnt inhibition also enhances thermogenesis in primary cells derived from brown adipose tissue but not epididymal adipose tissue (A) Dose-response curve of primary brown adipocytes treated with increasing dose of C59. RNA was extracted at Day 6 for qPCR analysis. Shown is the relative mRNA expression values normalized with housekeeping genes Rpl23. All data are presented as mean ± SEM by Student's t-test. *p < 0.05 comparing with control; ▴p < 0.05 comparing with 1nM. (B) Western Blot analysis on protein extracted at Day 6 from primary brown adipocytes with or without 10 nM C59 treatment. Also shown is protein extracted from brown adipose tissue (BAT), inguinal adipose tissue (WAT) and inguinal adipose tissue from mice underwent 7 d of 5°C cold exposure (WAT cold). (C) Primary brown adipocytes treated with 10 nM of C59 were harvested at 3 different time points (Day 2, Day 4 and Day 6) and subjected to qPCR analysis. (D) Western Blot analysis on protein extracted at Day2, Day4 and Day 6 from primary brown adipocytes during the course of differentiation treated with or without 10 nM C59 (E) Primary white adipocytes derived from epididymal fat pad were subjected to qPCR analysis. All data are presented as mean ± SEM by Student's t-test. *** p < 0.001 comparing with the control model. The fold of induction or inhibition is indicated when the change is significant and relevant.