Figures & data
Figure 1. Suv39h1/h2 expression is transiently induced during the early stage of 3T3-L1 adipogenesis. 3T3-L1 preadipocytes were induced to differentiation and cells were harvested at the time point indicated. Suv39h1/h2 mRNA (a) and protein (b) levels were measured by quantitative PCR analysis and immunoblotting, respectively. All data are expressed as mean ± SEM, n = 4
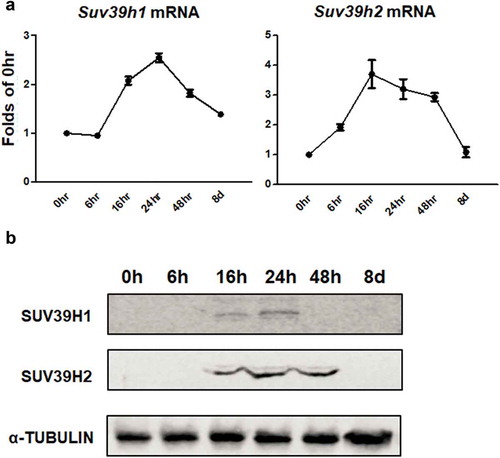
Figure 2. Suv39h1 or Suv39h2 knockdown suppresses 3T3-L1 adipogenesis. (a) Suv39h1 or Suv39h2 knockdown inhibits mRNA and protein levels of Suv39h1 or Suv39h2 in the 3T3-L1 cells. (b) Suv39h1 or h2 knockdown inhibits adipogenic gene expression in 3T3-L1 preadipocytes at day 4 of differentiation. (c) Oil Red O staining reveals less lipid accumulation in Suv39h1 orh2 knockdown cells at day 8 of differentiation. Microscopic image (upper) and whole well view (lower). 3T3-L1 preadipocytes were infected with Suv39h1 or h2 shRNA lentivirus, selected with puromycin and differentiated as described in the Methods. Gene expression was measured by quantitative PCR. All data are expressed as mean ± SEM, n = 4. *p < 0.05 vs. scramble control
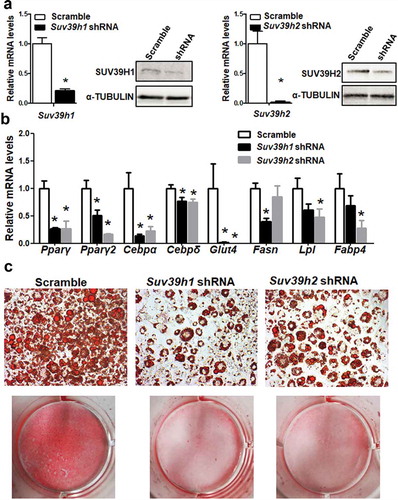
Figure 3. Suv39h1 knockdown inhibits adipogenic protein expression in 3T3-L1 preadipocytes. (a) Immunoblots of PPARγ and CEBPα protein. (b) Density quantitation of the immunoblots. 3T3-L1 preadipocytes were infected with Suv39h1 shRNA lentivirus, selected with puromycin and differentiated as described in the Methods. Protein levels were measured by immunoblotting. All data are expressed as mean ± SEM, n = 4. *p < 0.05 vs. scramble control
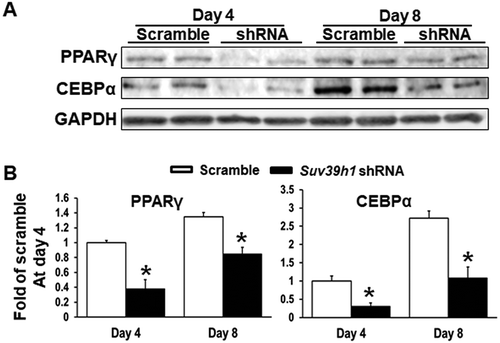
Figure 4. Suv39h1 overexpression promotes 3T3-L1 adipogenesis. (a) Suv39h1 overexpression promotes adipogenic gene expression in 3T3-L1 preadipocytes. (b) Oil Red O staining reveals more lipid accumulation in Suv39h1 overexpressing cells. 3T3-L1 preadipocytes were infected with the Suv39h1 pLVX lentivirus, selected with puromycin and differentiated as described in the Methods. Gene expression was measured by quantitative PCR. All data are expressed as mean ± SEM, n = 4. *p < 0.05 vs. scramble control
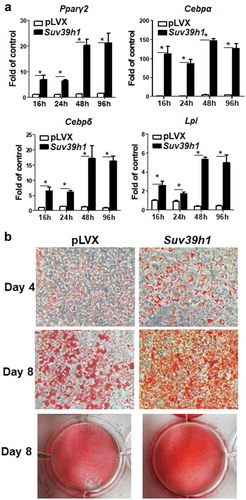
Figure 5. Wnt gene expression is reciprocally regulated by gain-or-loss of function of Suv39h1. The expression of three Wnt family members Wnt10a, Wnt10b and Wnt6 was significantly up-regulated in Suv39 h1 knockdown cells (a) while reciprocally down-regulated in Suv39 h1-overexpressing cells (b). 3T3-L1 preadipocytes were infected with Suv39h1 shRNA lentivirus or pLVX Suv39h1 expression lentivirus, selected with puromycin and differentiated as described in the Methods. Gene expression was measured by quantitative PCR. All data are expressed as mean ± SEM, n = 4. *p < 0.05 vs. scramble control or pLVX control
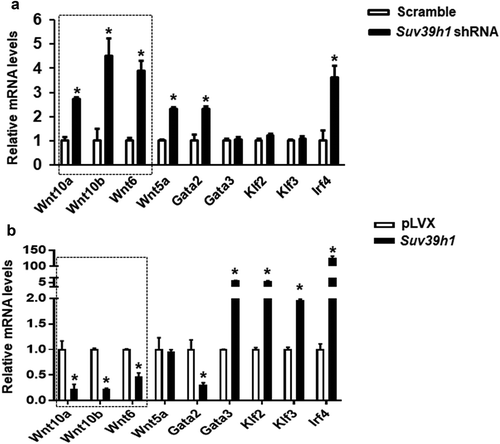
Figure 6. Suv39h1 regulates tri-methylated H3K9 (H3K9me3) at the Wnt10a promoter. Suv39h1 knockdown decreased (a), while Suv39h1 overexpression increased (b) H3K9me3 levels at the Wnt10a promoter in 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were infected with the Suv39h1 or h2 shRNA lentivirus, selected with puromycin and differentiated as described in the Methods. H3K9me3 at the Wnt10a promoter was assessed by ChIP assays with an anti-H3K9me3 antibody. All data are expressed as mean ± SEM, n = 3. *p < 0.05 vs. scramble control
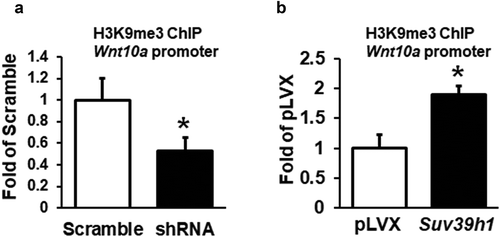
Figure 7. Wnt10a is a mediator of Suv39 h1’s action in adipogenesis. (a) Wnt10a knockdown substantially blocked the inhibitory effect of Suv39h1 deficiency in the expression of adipogenic markers. Suv39h1 knockdown preadipocytes were reversely transfected with Wnt10a siRNA and then differentiated into adipocytes as described in the Methods. Gene expression was measured by quantitative PCR. All data are expressed as mean ± SEM, n = 4. Bars labelled with different letters are statistically different from each other. (b) Oil Red O staining reveals increased lipid accumulation in Suv39 h1/Wnt10a double knockdown cells
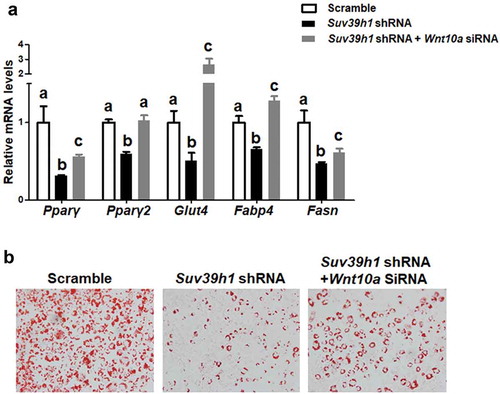
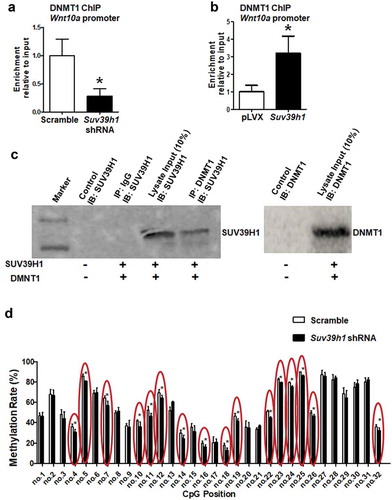
Figure 8. H3K9 methylation status affects recruitment of DNMT1 to the Wnt10a promoter. Suv39h1 knockdown decreased (a), while Suv39h1 overexpression (b) increased DNMT1 recruitment at the Wnt10a promoter in 3T3-L1 preadipocytes. 3T3-L1 preadipocytes were infected with Suv39 h1 shRNA lentivirus or overexpression lentivirus, selected with puromycin and differentiated as described in the Methods. DNMT1 binding to the Wnt10a promoter was assessed by ChIP assays with an anti-DNMT1 antibody. All data are expressed as mean ± SEM. *p < 0.05 vs. scramble control or pLVX control. (c) Immunoprecipitation of DNMT1 pulls down the SUV39 H1 protein. Immunoprecipitation and immunoblotting were conducted as described in the Methods. 10% Input: without IP, SUV39H1 (left blot) or DNMT1 (right blot) immunoblotting only. (d) Suv39h1 knockdown decreases CpG site methylation rate at the Wnt10a promoter. CpG methylation was measured by pyrosequencing analysis as described in the Methods. All data are expressed as mean ± SEM. *p < 0.05 vs. scramble control