Figures & data
Figure 1. Differentially expressed genes (DEGs) between the obesity and control cohorts. Volcano plot of GSE133099 and 11 substantially expressed genes were detected. Black, green, and red dots represent unchanged, downregulated, and upregulated genes, in that order
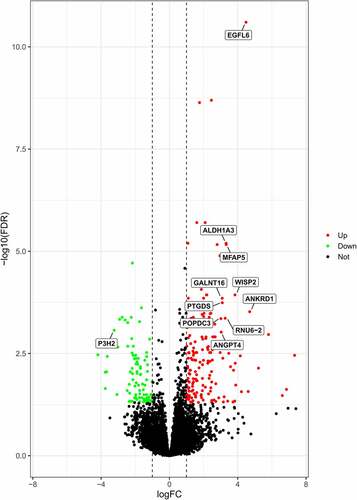
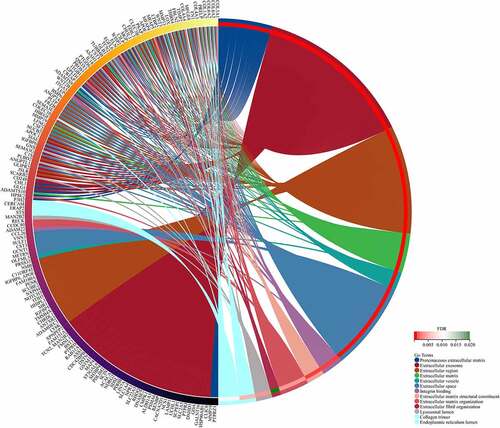
Table 1. Significantly enriched GO terms in obesity-related DEGs
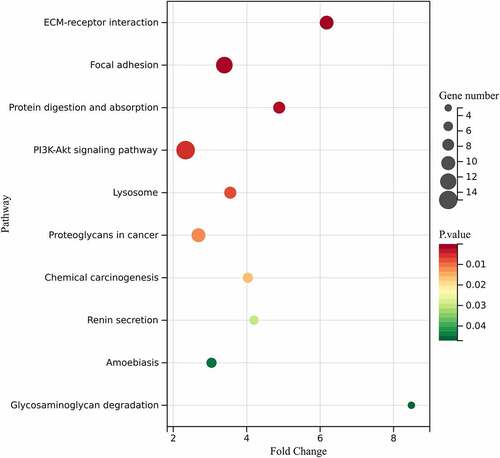
Table 2. Significantly enriched pathways in obesity-related DEGs
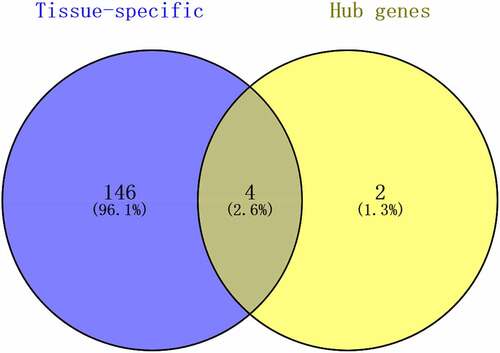
Table 3. Tissue-specific genes identified using BioGPS
Table 4. Nine FDA-approved drugs potentially targeting the four key genes