Figures & data
Table 1. HLA ligandome analysis and immunohistochemistry of OvCa samples.
Figure 1. Immunogenicity analysis of RMLPHAPGV-primed T cells. CD8+ T cells from healthy blood donors were primed in vitro using (A) aAPCs or (B+C) natural APCs. Numbers represent percentages of all T cells. (A) Exemplary tetramer staining of one donor primed with RMLPHAPGV-specific aAPCs. Tetramer staining was performed after three stimulations with aAPCs using control tetramer YLLPAIVHI and RMLPHAPGV tetramer. Numbers represent percentage of all T cells. This figure is representative of nine donors tested positive for RMLPHAPGV-specific T cells after in vitro expansion with aAPCs. (B) Exemplary intracellular IFNγ staining of one donor primed with RMLPHAPGV-presenting natural APCs. Staining was performed after five stimulations with natural APCs using control peptide YLLPAIVHI and RMLPHAPGV peptide. (C) T cells, which have been primed in vitro using natural APCs, (from panel B) were stained with irrelevant YLLPAIVHI and RMLPHAPGV tetramer.
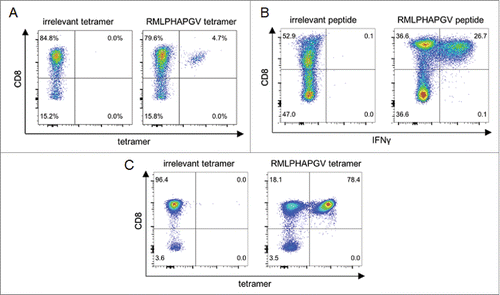
Table 2. Immunogenicity of HDAC-derived HLA ligands.
Figure 2. Multi-functionality and cytotoxicity of in vitro primed T cells reacting to RMLPHAPGV. (A) T cells show significant expression of TNFα, MIP-1β, IL-2 and CD107a after 6 h incubation with RMLPHAPGV. Numbers in the plot reflect percentage of reactive T cells for the respective marker. (B) Reactive T cells show different degrees of functionality. Only T cells expressing at least one activation marker (60.1% of all RMLPHAPGV-specific T cells) are displayed. 39.9% of RMLPHAPGV-tetramer-specific T cells did not react upon peptide stimulation. (C) Different combinations of activation markers are represented within the reactive T-cell population. (D) Percentage of specific lysis by antigen-specific T cells was determined by 51chromium release assay after 24 h coincubation with different RMLPHAPGV peptide-loaded or peptide-unloaded OvCa cell lines or HLA-negative K562 cells. Experiments were performed in triplicates with a 30:1 E:T ratio.
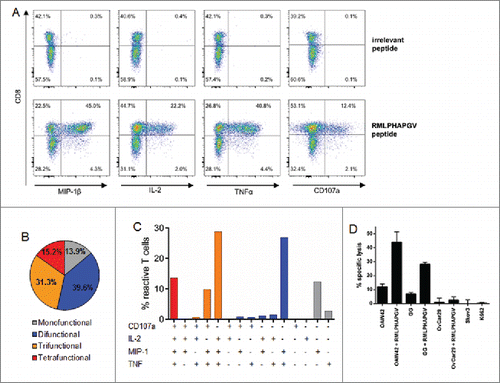
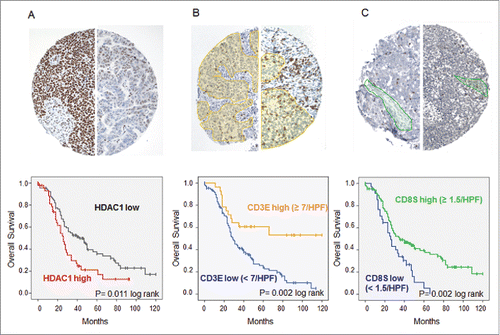
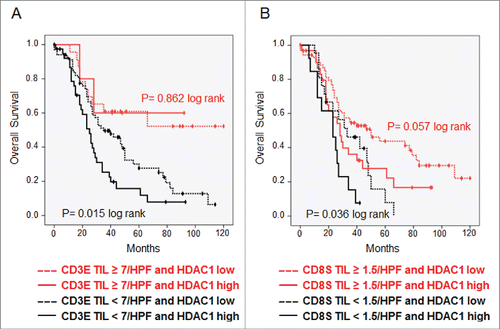
Figure 3. Immunohistochemistry of high-grade serous OvCa. Immunohistochemical stainings on TMAs with 136 high-grade serous OvCa samples were performed. (A) Expression of HDAC1: Top: Immunohistochemical staining for HDAC1 of two exemplary OvCa samples with high HDAC1 expression (score 12, left) and low HDAC1 expression (score 3, right). Bottom: Impact of HDAC1 on OS of high-grade serous OvCa stage III and IV (Kaplan Meier analysis, low expression: score < 9, n = 94; high expression score ≥ 9, n = 45). (B) Intraepithelial CD3+ TILs: Top: Immunohistochemical staining for CD3+ TILs of two exemplary OvCa samples with lower infiltration (left) and higher infiltration (right) of TILs (epithelial compartment is highlighted in yellow). Bottom: Impact of epithelial CD3+ TILs on OS of high-grade serous OvCa stage II to IV (Kaplan Meier analysis, low: < 7 cells/HPF, n = 108; high: ≥ 7 cells/HPF, n = 28). (C) Intrastromal CD8+ TILs: Top: Immunohistochemical staining for CD8+ TILs of two exemplary OvCa samples with lower infiltration (left) and higher infiltration (right) of TILs (stromal compartment is highlighted in green). Bottom: Impact of stromal CD8+ TILs on OS of high-grade serous OvCa stage II to IV (Kaplan Meier analysis, low: < 1.5 cells/HPF, n = 91; high: ≥ 1.5 cells/HPF, n = 45).