Figures & data
Figure 1. Immunochistochemical staining of CD19 and PD-L1 in IBCa and fibroadenoma tissue of breast. Sections of fibroadenoma tissues and IBCa were stained with anti-CD19, anti-PD-1 and anti-PD-L1 antibodies, as described in “Materials and Methods”, shown in are representative microphotographs (× 400). (A) The density of CD19+ B lymphocytes is significantly higher in IBCa tissue than that in fibroadenomas of breast. (B) The density of CD19+ B lymphocytes in grade 1, 2 and 3 IBCa. C: PD-L1 is strongly expressed in the cell membrane, cytoplasm or both the IBCa tissues than that in fibroadenomas of the breast. Isotype-matched mouse IgG1 mAb was used as control.
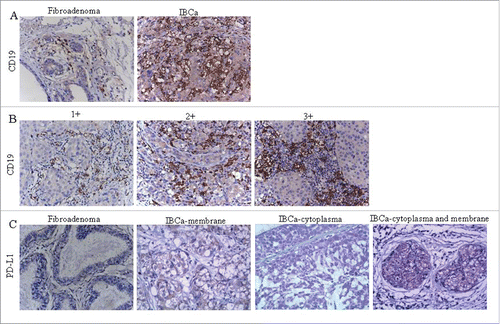
Figure 2. The colocalization of CD19 and IL-10 expression in IBCa tissues. The colocalization of CD19 and IL-10 expression in IBCa were analyzed by immunohistochemistry and immunoflurescence staining as described in “Materials and Methods”, shown in are representative microphotographs (× 400). (A) The immunohistochemistry staining of serial sections showed the location of CD19 expression as well as IL-10 in TILs of IBCa tissue. (B) Confocal Laser-Scanning Microscopy was applied to detect CD19 and IL-10 in IBCa specimens. Membrane of B cells was visualized by CD19 staining (Green), cytoplasm are stained with IL-10 (Red), and nuclei are counterstained with DAPI (Blue). Merged figure exhibit a colocalization of CD19 and IL-10 in the B cells.
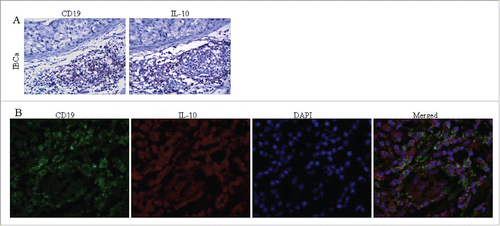
Table 1. Comparisons of the frequency of tumor-infiltrating B cells in breast fibroadenoma and IBCa tissue.
Table 2. Relationship between the tumor-infiltrating CD19+ B cell density and the clinicopathological parameters of IBCa patients.
Table 3. PD-L1 expression in breast fibroadenoma and IBCa tissue.
Table 4. Relationship between PD-L1 expression in IBCa tissue with histopathological features of IBCa.
Table 5. Relationship between IL-10 expression in IBCa tissue with histopathological features of IBCa.
Table 6. Correlation of tumor-infiltrating CD19+ B cells with PD-L1 expression in cell membrane and cytoplasm of IBCa tissue.
Table 7. Correlation of tumor-infiltrating CD19+ B cells with IL-10 expression in IBCa tissue.
Figure 3. PD-L1 mediates the differentiation of CD19+ B cells into CD19+CD24+CD38+ B cell subtype. (A) The expression of PD-L1 and PD-1 on breast cancer cell lines was analyzed by flow cytometry. MDA-MB231 expresses high level of PD-L1 (94.7%) compared with MCF-7 (17.3%); Both MCF-7 and MDA-MB231 lacks PD-1 expression. (B) CD19+ B lymphocytes/ breast cancer cells co-culture systems were established to investigate the interaction between PD-L1 expression in breast cancer cells and CD19+ B cells. 16.1% of CD19+CD38+CD24+ B cells were detected in PD-L1hi MDA-MB231, while only 9.38% of CD19+CD38+CD24+ B cells were detected in PD-L1lo MCF-7. (C) To confirmed the relationship between PD-L1 and CD19+ B cells, PD-L1siRNA were used to knockdown the expression of PD-L1. (D) Co-culture of CD19+ B cells and PD-L1hi MDA-MB231 with or without siRNA. The percentage of CD19+CD38+CD24+ B cells were reduced when CD19+ B cells were co-cultured with MDA-MB231 cells with PD-L1 knocked down. (E) A high level of IL10 was detected in PD-L1hi co-culture compared to that in PD-L1lo co-culture system (P < 0.05). And CD19+CD24+CD38+ B cells produced IL-10 following activation by PMA and ionomycin.
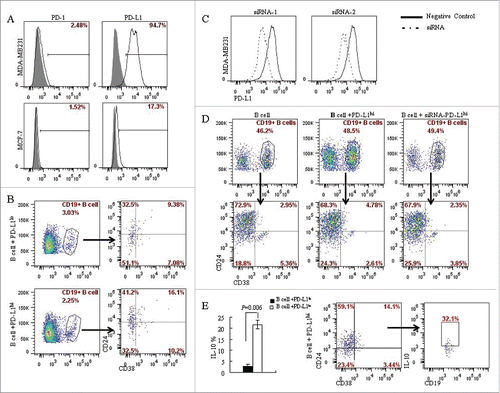
Figure 4. Phenotype and function of CD19+ B cells from IBCa patients. To investigate the phenotype and function of CD19+ B cells in patients and healthy individuals, CD19+ B cells were isolated from peripheral blood or breast tumor tissues, incubated with CD19-ECD, CD38-APC and CD24-PE-cy7, and analyzed by flow cytometry. CD4+ T cells in patients and healthy individuals were purified and incubated with CD4-ECD, CD25-PE-cy7, fixed and stained by Foxp3-APC. B/T lymphocytes co-culture systems were set up to examine the interaction between CD19+ B cells and CD4+ T cells. (A) Higher percentage of CD19+CD38+CD24+ B cells were detected in fresh IBCa tissues that in breast fibroadenoma tissue (6.38% vs. 2.79%). (B) Significantly higher level of CD19+CD38+CD24+ B cells was detected in the PBMC of IBCa patients compared with that in fibroadenoma or health individuals (p = 0.0176). (C) Elevated expression of IL-10 (36.4%) was detected in CD19+CD24+CD38+ B cells activated by LPS, PMA and ionomycin compared to that in CD19+CD24+CD38− B cells (2.90%, P < 0.05). (D) A total of 80.4% of CD4+CD25+Foxp3+ T cells were found in the co-culture system, in which both CD19+ B cells and CD4+ T cells were originated from IBCa patients, versus 30.8% of CD4+CD25+Foxp3+ T cells in co-culture system of CD19+ B cells from patients and CD4+ T cells from health individuals. However, only 7.53% and 8.13% of CD4+CD25+Foxp3+ T cells were detected in the co-culture system of CD19+ B cells from health individuals with CD4+ T cells originated from health individual or IBCa patients, respectively. (E) Significantly higher level of CD4+CD25+Foxp3+ Treg cells were detected in the PBMC of IBCa patients compared with that in fibroadenoma patients or health individuals (P < 0.0001).
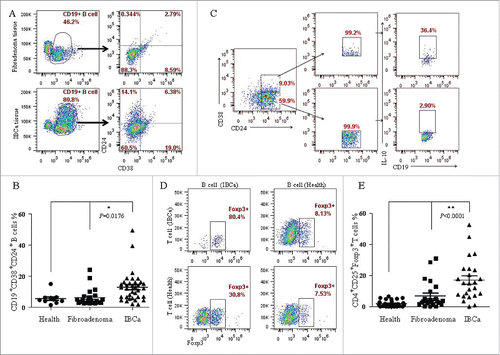
Figure 5. PD-L1 contributed to the induction of the CD4+CD25+Foxp3+ Tregs by CD19+ B cells in in vitro system. The potential role of PD-L1 in the B cells and T cells interaction were examined in co-culture experiments of CD4+ T cell with CD19+ B cells, or CD19+ B cells pre-cultured with MCF-7 or MD-MB231 cells, or MD-MB231 cells with or without PD-L1 knocked down. CD4+ T cells were isolated by CD4+ positive selection kit from peripheral blood of healthy individuals. Following activation with anti-human CD3 and anti-human CD28 antibodies, CD4+ T cells and B cells as described above were mixed together at a ratio of 1:1, and analyzed by flow cytometry following culture for the specified time as described in the Materials and Methods section. (A) A total of 48.6% CD4+CD25+ Foxp3+ T cells were produced in co-culture system in which B cells were pre-cultured (or stimulated) by PD-L1hi MD-MB231, 21.8% CD4+CD25+ Foxp3+ T cells in co-culture system of T cells with B cells stimulated by PD-L1lo MCF-7. (B) A total of 5.09% and 12.5% of CD4+CD25+ Foxp3+ T cells were detected in co-culture with B cells stimulated by MD-MB231 with or without PD-L1 depletion by PD-L1-siRNA or PD-L1-NC (negative control), respectively.

