Figures & data
Figure 1. Acceleration of tumor development associated with NOD genetic background. Mice transgenic for the RET oncogene were monitored weekly for the onset of tumors at the eye, the primary site (A) and for local (B) and distant (C) cutaneous (cut.) metastases. The comparison included mice with a B6 background, F1 mice intercrossed with NOD mice, and these F1 mice backcrossed with NOD at the second (F2), third (F3) and tenth (F10) generation. Data are displayed using the Kaplan–Meier representation.
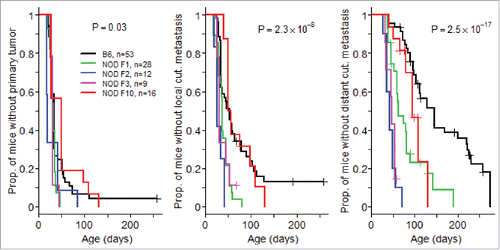
Figure 2. Alteration of T cell subsets in (NODxB6)F1 and NOD F5-F10 backcrossed mice carrying the RET transgene. (A) Density of CD4+ and CD8+ T cells in tumors. Boxplots show the mean and interquartile range (box) and the 5–95 percentiles (whiskers). B6, n = 10; (NODXB6)F1, n = 10; NOD F5-F10, n = 13. (B, C) The proportion of CD4+ T cells expressing Foxp3 (B), and of CD8+ T cells expressing IFNγ (C) was determined by intra-cellular immunolabeling and flow cytometry analysis in the peripheral (axillary and inguinal) lymph nodes (LN), submandibular lymph nodes (SM) draining the facial tumors, the spleen (S) and the tumor (T) of RET-transgenic mice and their non-transgenic littermates. Expression of IFNγ was assessed after stimulation with PMA and ionomycin for 4 h. The average proportions in the various conditions were compared with pairwise t tests. The resulting p values were systematically corrected for multiple testing using the Bonferroni method. For clarity, only relevant significant p values are reported. (D) Correlation between the proportions of CD4+Foxp3+ T cells and of CD8+ T cells expressing IFNγ in F1 hybrid (open symbols, dashed regression line) and F5–F10 backcrossed (closed symbols, solid regression line) mice. (E) Correlations of clinical score with proportion of CD4+Foxp3+ T cells in spleens (open symbols, dashed regression line) or in tumors (close symbols, solid regression line) in F1 and F5–F10 backcrossed mice. All correlations were tested using the non-parametric Spearman method. Number of samples in each group: B6.RET− LN, n = 3; B6.RET+ LN, n = 7; B6.RET+ SM, n = 5, B6.RET− S, n = 3; B6.RET+ S, n = 10; B6.RET+ T, n = 10; (NODxB6)F1.RET− LN, n = 7; (NODxB6)F1.RET+ LN, n = 6; (NODxB6)F1.RET+ SM, n = 6; (NODxB6)F1.RET− S, n = 10; (NODxB6)F1.RET+ S, n = 13; (NODxB6)F1.RET+ T, n = 9; NOD F5–F10.RET− LN, n = 22; NOD F5–F10.RET+ LN, n = 16; NOD F5–F10.RET+ SM, n = 16; NOD F5–F10.RET− S, n = 15; NOD F5–F10.RET+ S, n = 13; NOD F5–F10.RET+ T, n = 11.
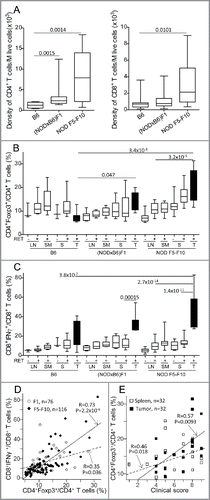
Figure 3. Marked alteration of Dectin-1 and Trem-1 expression on myeloid cells. Proportions of CD11b+Ly6G− (A) and CD11b+Ly6G+ (B) cells among CD45+ cells in spleens (S) and tumors (T) were determined by immunolabeling and flow cytometry. Expression of Dectin-1 (C–F) and Trem-1 (G, H) was determined on CD11b+Ly6G+ (C–E, G) and CD11b+Ly6G− (F, H). Data are depicted as the proportion of positively labeled cells (A–C, F–H) or using the mean fluorescence intensity (MFI) of the labeling (D, G). Representative histograms of fluorescence intensities are also shown for Dectin-1 on CD11b+Ly6G+ cells (E). Bars in scatterplots indicate the mean sample and its standard error. The sample means in the various conditions were compared with pairwise t tests. The resulting p values were corrected using the Bonferroni method. For clarity, only relevant significant p values are reported. Number of samples per group: B6.RET− S, n = 8; B6.RET+ S, n = 17; B6.RET+ T, n = 12; (NODxB6)F1.RET− S, n = 11; (NODxB6)F1.RET+ S, n = 16; (NODxB6)F1.RET+ T, n = 10; NOD F5–F10.RET− S, n = 55; NOD F5–F10.RET+ S, n = 67; NOD F5–F10.RET+ T, n = 17.
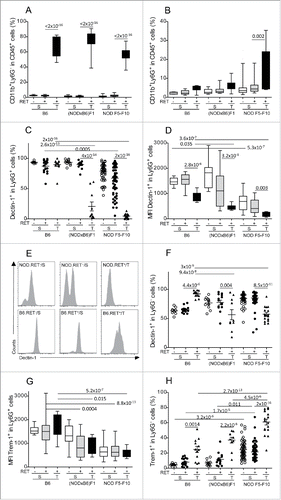
Figure 4. Correlation between myeloid subsets, including CD11b+Ly6G+Dectin-1+ (upper panels) and CD11b+Ly6G−Trem-1+ (lower panels) with T lymphoid subsets, including CD4+Foxp3+ (left panels) and CD8+IFNγ+ (right panels) in 36 tumors from (NODxB6)F1 and NOD F5–F10 mice. Significance of correlations was assessed with the non-parametric method of Spearman.
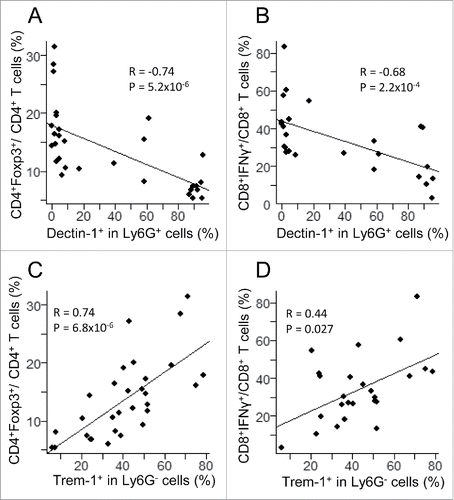
Figure 5. Major changes in subsets of myeloid cells are associated with the NOD genetic background. Spleen (S) and tumor (T) CD11b+Ly6G+ (A) and CD11b+Ly6G− (B) cells were gated electronically and the percentages of cell subsets defined by Dectin-1/Trem-1 double labeling were determined and are depicted using pie charts. Dec, Dectin-1; Trm, Trem-1.
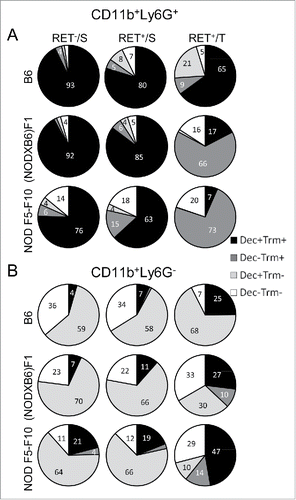
Figure 6. Protection against tumor development by curdlan treatment. (A) (NODxB6)F1.RET+ transgenic mice (6–7 weeks of age) were injected intraperitoneally twice with curdlan (100 µg/g of body weight, n = 12) or with PBS (n = 9) with a one-week interval and were monitored weekly for tumor progression. The data are pooled from two experiments conducted at an interval of 6 mo. The mean tumor score and its standard deviation are shown at each timepoint. The difference of tumor scores between both groups was tested using a linear model with mixed effects. (B) Reduction of CD4+Foxp3+ T cells in tumors of curdlan-treated mice.
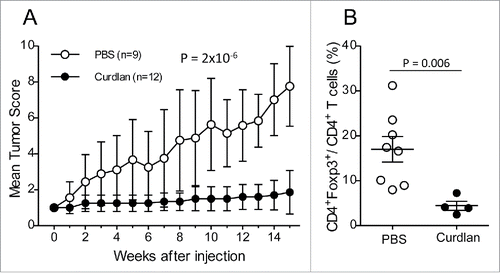
Figure 7. Less severe tumor evolution and upregulation of Dectin-1 expression in (NODxB6)F1.RET+.Nos2−/− mice. Hybrid (NODxB6)F1.RET+ transgenic mice proficient (open circles, n = 24) or deficient (dark circles, n = 27) for the Nos2 gene were monitored for tumor development every other week for 7 mo. The mean tumor score and its standard deviation are shown at each timepoint. The difference of tumor scores between both groups was tested using a linear model with mixed effects. (B, C) Upregulation of Dectin-1 expression on CD11b+Ly6G+ (B) and CD11b+Ly6G− (C) myeloid cells in spleen (S) and tumors (T) of (NODxB6)F1 and B6.RET+ mice deficient for Nos2 compared with their Nos2+/+ counterparts. (D, E) Dectin-1/Trem-1 double labeling of myeloid subsets in spleens and tumors. CD11b+Ly6G+ (D) and CD11b+Ly6G− (E) cells were gated electronically and the percentages of cell subsets defined by Dectin-1/Trem-1 double labeling were determined in mice with indicated genetic background and are depicted using pie charts. Dec, Dectin-1; Trm, Trem-1. (F) Quantification by qPCR of RET transgene expression in tumors (T) and in lymphoid organs, including spleen (S), peripheral lymph nodes (LN) and submandibular lymph nodes (SM), of RET+ mice with indicated genetic background. Normalized relative quantities (NRQ) of RET messages were determined as described in Materials and Methods. Number of samples per group: B6.RET− S, n = 8; B6.RET+ S, n = 17; B6.RET+ T, n = 12; B6.Nos2−/−.RET− S, n = 7; B6.Nos2−/−.RET+ S, n = 12; F1.Nos2−/−.RET− S, n = 17; F1.Nos2−/−.RET+ S, n = 16; F1.Nos2−/−.RET+ T, n = 4; F1.RET− S, n = 11; F1.RET+ S, n = 14; F1.RET+ T, n = 10.
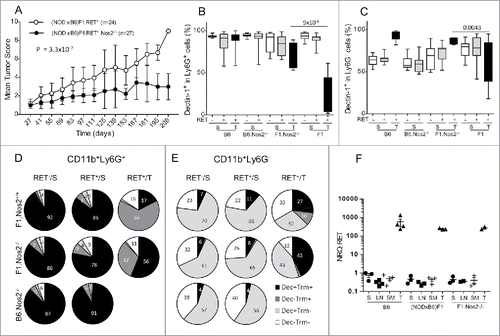
Figure 8. Gating strategy for flow cytometry labeling of spleen myeloid cells from an NOD F5–F10 RET+ mouse. A sample with a decreased level of Dectin-1 expression was selected. Figures indicate the percentages of events in the gates or in each quadrant.
Table 1. Primer pairs used for real-time PCR experiments.
