Figures & data
Figure 1. Immunohistochemical staining of ICOS in colorectal cancer. Tumor cores of CRC samples were stained with an ICOS specific monoclonal antibody (clone SP98) by immunohistochemistry. Representative images of ICOS+++ (A), ICOS++ (B), ICOS+ (C) and ICOS-(D).
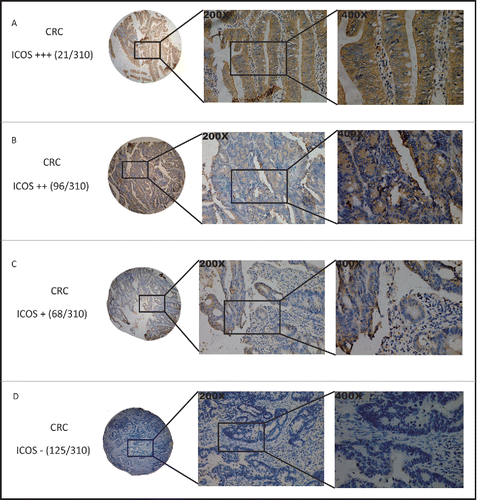
Figure 2. Expression of ICOS is associated with metastasis and other pathological features of CRC patients. The scores of ICOS staining in individual CRC punches (n = 310) were correlated with different status of lymphatic metastasis (A), distant metastasis (B), TNM stage (C) and CEA level (D).
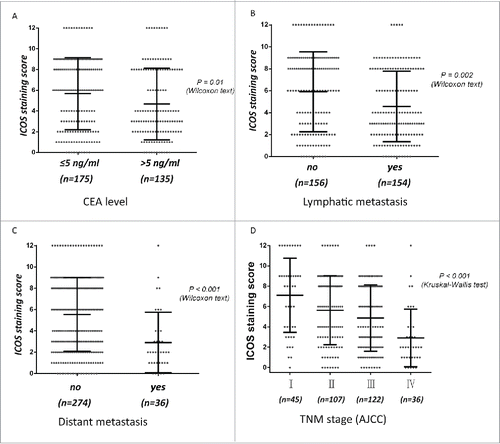
Table 1. Correlations between ICOS expression and clinicopathologic features in 310 colorectal cancer patients.
Figure 3. Kaplan–Meier analysis of overall survival in colorectal cancer patients and differences were analyzed by Log Rank test. (A, B), High expression of ICOS is associated with a good overall survival (OS) (p < 0.001) and a longer DFS (p < 0.001). ICOS was detected by monoclonal antibody (ab105227, abcam).
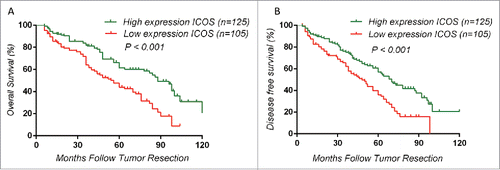
Table 2. Univariate and multivariate analyses of prognostic parameters for survival in 230 colorectal cancer patients.
Figure 4. ICOS expression pattern in tumor sites and peripheral blood. (A) CRC surgical tumor tissues were enzymatically digested and stained with fluorochrome labeled Abs, followed by flow cytometry analysis. (i) Representative figures of ICOS expression on CD4+ and CD8+ T cells, analyzed by flow cytometry, in tumor-infiltrated leukocytes. (ii) Quantitation of the percentage of ICOS+CD4+ or ICOS+CD8+ T cells among all of the CD4+ or CD8+ T cells gotten from tumor-infiltrated leukocytes, respectively (n = 26). (B) The similar protocol is as (A), except for the specimens from pericarcinous tissues (i) or distal normal tissues (ii). (C) The similar protocol is as (A), except for the specimens from peripheral blood. (D) CRC samples were stained with ICOS antibody by immunohistochemistry. At the same time, the samples were enzymatically digested to get the TILs and ICOS expression on CD4+ T cells were analyzed by flow cytometry. (E) Correlation between percentage of ICOS+ among CD4+ T cells and ICOS staining scores in the CRC tumor tissue (n = 13). Data are presented as averages ± SEM, statistical difference was evaluated by Student's t test. **p < 0.01; ***p < 0.001.
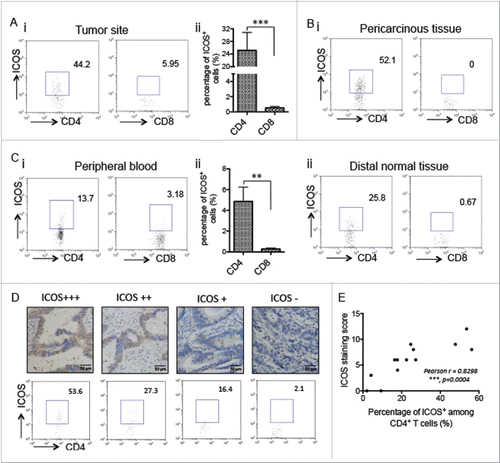
Figure 5. ICOSL expression pattern in tumor sites. CRC surgical tumor tissues were enzymatically digested and stained with fluorochrome labeled Abs, followed by flow cytometry analysis. (A) ICOSL expression in macrophages (CD11b+CD14+CD16−). (B) ICOSL expression in DCs (CD11c+MHC-II+). (C) ICOSL expression in B cells (CD19+CD3−). (D) ICOSL expression in tumor cells (EpCAM+CD45−). (E) Quantitation of the percentage of ICOSL+ cells among macrophages, DCs, B cells and tumor cells gotten from tumor-infiltrated leukocytes, respectively (n = 26). Data are presented as averages ± SEM, statistical difference was evaluated by Student's t test. **p < 0.01; ***p < 0.001.
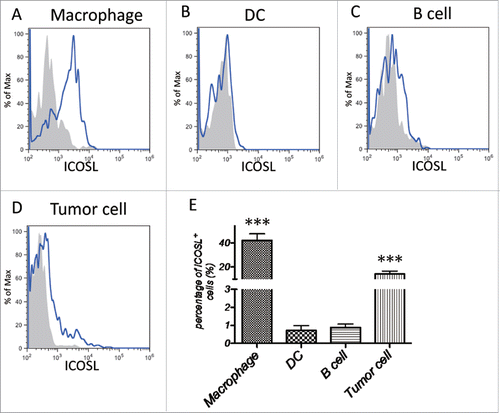
Figure 6. The expression of ICOS is associated with other immune checkpoints and immune cells. (A–C) Correlation between the percentage of ICOS+ T cells and that of CD28+ T cells (A), CTLA4+ T cells (B) and PD-1+ T cells (C) among total tumor-infiltrated leukocytes. (D–G) Correlation between the percentage of ICOS+ T cells and that of CD8+ T cells (D), CD4+ T cells (E), CD4+CD25+ T cells (F) and macrophages (CD11b+CD14+CD16−) (G) among tumor-infiltrated leukocytes.
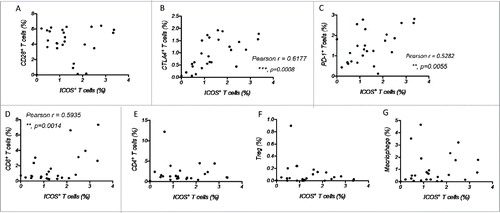
Figure 7. Characterization of tumor-infiltrating ICOS+CD4+ T cells, (A) CD45+ cells were enriched from primary CRC tumor tissues and the ICOS+CD4+ and ICOS−CD4+ T cells were subsequently sorted by a cell sort. (B) Relative mRNA level of transcriptional factors T-bet (Th1), GATA3 (Th2), RORγt (Th17) and FoxP3 (Treg) of ICOS+CD4+ and ICOS−CD4+ T cells were determined by realtime-PCR. (C) Protein level of cytokines IFNγ (Th1), IL4 (Th2), IL17 (Th17) and IL10 (Treg) in the culture medium of ICOS+CD4+ and ICOS−CD4+ T cells activated by leukocyte activation cocktail for 18 h were examined by ELISA. Data are presented as averages ± SEM, statistical difference was evaluated by Student's t test. *p < 0.05; **p < 0.01.
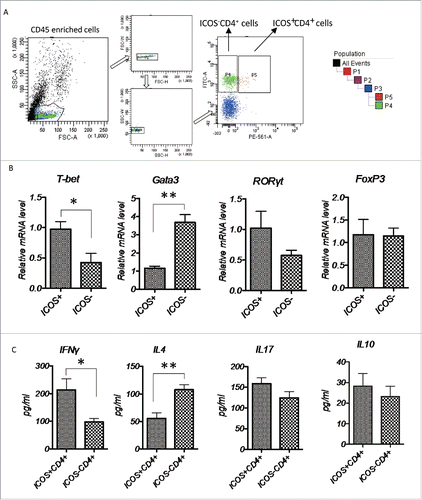
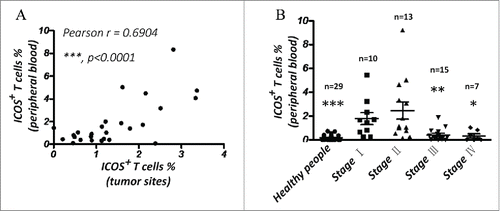
Figure 8. The percentage of ICOS+ T cells in PBMC is a good marker for prognosis of CRC. (A) Correlation between percentage of ICOS+ T cells in peripheral blood and that in primary tumor tissues. (B) Quantity of ICOS+ T cells in peripheral blood of luminal breast cancer patients by flow cytometry. Data are presented as averages ± SEM, statistical difference was evaluated by Student's t test. *p < 0.05; **p < 0.01; ***p < 0.001.