Figures & data
Figure 1. ApoAI mimetic peptide L37pA potentiates the antiviral and cytotoxic activity of IFNα. (A) Cytopathic effect reduction assay in L929 cells pretreated overnight with 2-fold serial dilutions of IFNα starting from 125 U/mL with L37pA (200 µg/mL), L37mut (200 µg/mL) or with no peptide. Cells viability was quantified 24 h after EMCV infection. (Extra sum-of-squares F test ***p < 0.001). (B) Cytotoxicity assay in L929 cells incubated for 3 d with IFNα (1500 U/mL) and L37pA (200 µg/mL), L37mut or with no peptide (one way ANOVA followed by Dunnett's multiple comparison test. ***p < 0.001). (C) Quantitative real time PCR analysis of interferon-stimulated genes after 3 h of stimulation of L929 cells with IFNα (200 U/mL), L37pA (200 µg/mL) or the combination. Data are expressed as mean + SEM (one way ANOVA, followed by Dunnett's multiple comparison test. **p < 0.01, ***p < 0.001).
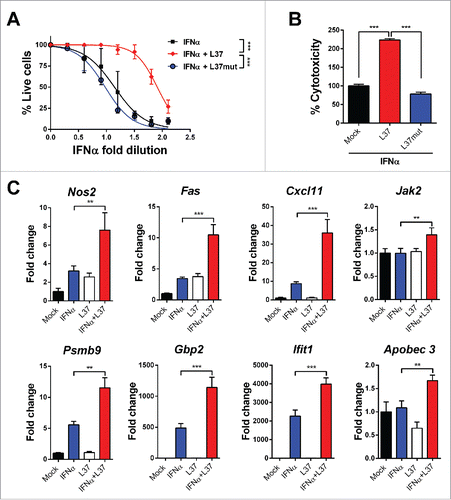
Figure 2. Mechanisms of IFNα and L37pA synergy. We determined the expression of Cxcl11 and Fas as readout of the effect of IFNα plus L37pA using quantitative real time RT-PCR in L929 cells treated as follows: (A) Cells were stimulated with IFNα (200 U/mL) for 3 h alone or in combination with L37pA (200 µg/mL), high density lipoprotein (HDL) (5 µg/mL), apolipoprotein A-I (ApoA-I) (30 µg/mL) or serum amyloid A (SAA) (30 µg/mL). (B) Cells were stimulated with IFNα (200 U/mL) for 3 h in combination with L37PA (200 µg/mL), the Alzheimer amyloid β peptide (Aβ) (200 µg/mL), cathelicidin (LL37) (200 µg/mL), phenol-soluble modulin 1 (M1) (200 µg/mL) or LPS (160 µg/mL). (C) Cells were pretreated with neutralizing antibodies against TLR2 (5 µg/mL), TLR4 (40 µg/mL) or with the combination for 1 h. Then, cells were treated with IFNα (200 U/mL) alone or plus L37pA (200 µg/mL) for 3 h. (D) Cytotoxicity assay in L929 cells incubated for 3 d with IFNα (1500 U/mL) and L37pA (200 µg/mL), and pretreated with neutralizing antibodies against TLR2 (5 µg/mL), TLR4 (5 µg/mL) for 1 h. Data are expressed as mean + SEM (one way ANOVA, followed by Dunnett's multiple comparison test. **p < 0.01, ***p < 0.001).
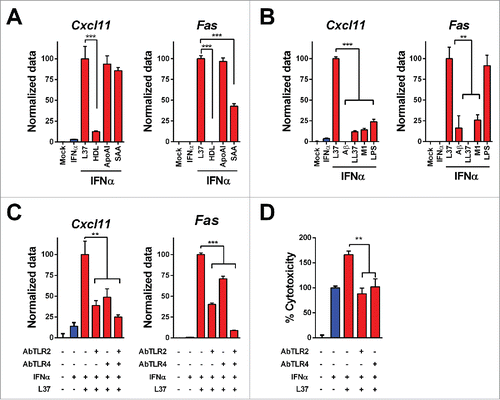
Figure 3. Co-expression of L37pA and IFNα eliminates AAV genomes in the liver. C57BL/6 mice were treated with 1 × 1012 vg AAV-Luc (n = 6), 5 × 1011 AAV-Luc and 5 × 1011 AAV-IFN (n = 6) or 5 × 1011 AAV-L37 and 5 × 1011 AAV-IFN (n = 6). Four weeks after virus administration, we determined the following parameters: (A) IFNα serum levels were measured by ELISA. (B) White blood cells (WBC), platelets (PLT) and red blood cells (RBC). (C) Viral genomes in the liver were determined by quantitative real time PCR. (D) ISGs expression in the liver. Data are expressed as mean + SEM (one way ANOVA, followed by Bonferroni's multiple comparison test. **p < 0.01, ***p < 0.001). (E) Kaplan–Meier plot representing the survival of C57BL/6 mice treated with 5 × 1011 AAV-Luc and 5 × 1011 AAV-IFN (n = 6) and the C57BL/6 mice (n = 6), Rag1 knockout mice (n = 5), Rag2γc knockout mice (n = 7), SR-B1 knockout mice (n = 3) treated with 5 × 1011 AAV-L37 and 5 × 1011 AAV-IFN. (Log rank test. ***p < 0.001). (F) Viral genomes in the liver were determined by quantitative real time PCR in mice that survived. Data is normalized to the viral genomes of wild type mice treated with 5 × 1011 AAV-Luc and 5 × 1011 AAV-L37 (one way ANOVA, followed by Dunnett's multiple comparison test. **p < 0.01). Results were confirmed with at least two independent experiments.
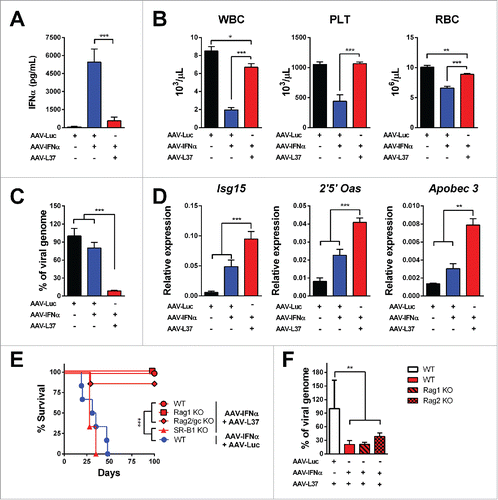
Figure 4. Antitumor effect in mice treated with AAV-IFN and AAV-L37. C57BL/6 mice were treated with 1 × 1012 vg AAV-Luc (n = 6), 5 × 1011 AAV-Luc and 5 × 1011 AAV-IFN (n = 6) or 5 × 1011 AAV-L37 and 5 × 1011 AAV-IFN (n = 12). (A) Experimental protocol. (B) 21 d after tumor inoculation, presence of tumors in the liver was assessed by echography and the percentage of tumor bearing animals is represented. (C) Survival is represented with a Kaplan–Meier plot (Log rank test. **p < 0.01). Results were confirmed with at least two independent experiments.
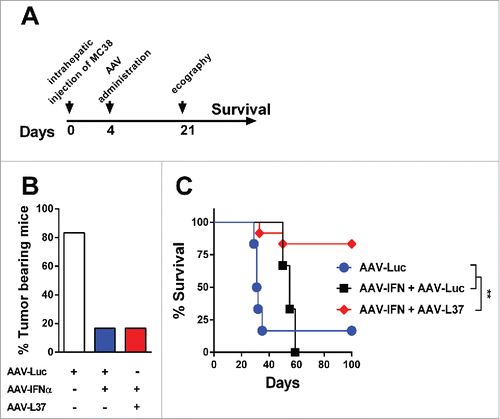
Figure 5. Inhibition of SR-B1 impairs IFNα function. L929 cells were transfected with siRNA targeting SR-B1 (siSR-B1) or control scrambled siRNA (siCTL) for 48 h. (A) SR-B1 mRNA was determined by quantitative real time PCR. Data are expressed as mean + SEM. (t-test. ***p < 0.001). Total SR-B1 protein was detected by immunoblot and β-actin is shown as loading control. (B) The silenced cells were stimulated with IFNα (200 U/mL) for 2 h. Then, ISGs expression was determined using quantitative real time PCR. mRNA levels were normalized to Rplpo. (C) Quantitative real time PCR analysis of ISGs expression after 3 h of stimulation with IFNα (200 U/mL) in L929 cells pretreated for 1 h with BLT1 (15 µM), ITX-5061 (30 µM) and Glyburide (500 µM). (D) Flow cytometry analysis of the IFNα receptor 1 (IFNAR1) in L929 cells treated with BLT-1 (15 µM) for 3 h. Normalized geometric mean of three samples (left panel) and representative histogram (right panel). Data are expressed as mean + SEM (E) Phosphorylated STAT1 and total STAT1 protein determined by immunoblotting in L929 cells stimulated with IFNα (1500 U/mL) for 30 min after incubation of BLT-1 (15 µM) or DMSO for 1 h. (F) Cytopathic effect reduction assay in mouse L929 cells pretreated overnight with 2-fold serial dilutions of IFNα starting from 125 U/mL in the presence or absence of BLT-1 (15 µM). Cells viability was quantified 24 h after EMCV infection (Extra sum-of-squares F test ***p < 0.001). (G) Cytotoxicity assay in mouse L929 incubated for 3 d with IFNα (1500 U/mL) in the presence or absence of BLT-1 (15 µM). (t-test or one way ANOVA, followed by the Dunnett's Multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001).
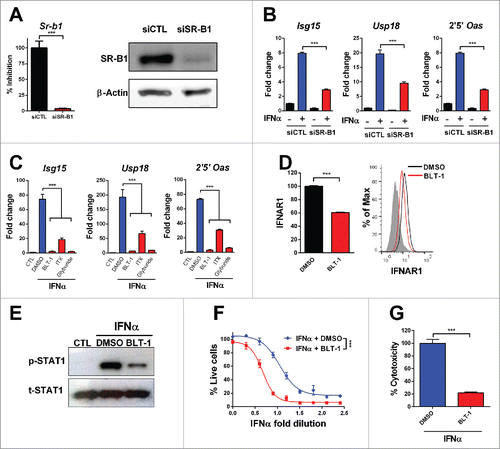
Figure 6. SR-B1 is necessary for the clathrin-mediated endocytosis. (A) Quantitative real time PCR analysis of ISGs expression 3 h after stimulation with IFNα (200 U/mL) in L929 cell pretreated for 1 h with chlorpromazine (CPZ) (10 µM) or BLT1 (15 µM). Data are expressed as mean + SEM (t-test. *p < 0.05). (B) Flow cytometry analysis of the IFNα receptor 1 (IFNAR1) prior treatment with BLT-1 (15 µM) or CPZ (10 µM) for 3 h and flow cytometry analysis in L929 cells treated with BSA-FITC (C) or L37pA-FITC (D) for 1 h prior treatment with CPZ (10 µM) or BLT-1 (15 µM) for 30 min at 37°C or 4°C. The normalized geometric mean (GM) fluorescence (number of replicates = 3) and a representing image at 37°C is shown. Data are expressed as mean + SEM (one way ANOVA, followed by Dunnett's multiple comparison test. ***p < 0.001).
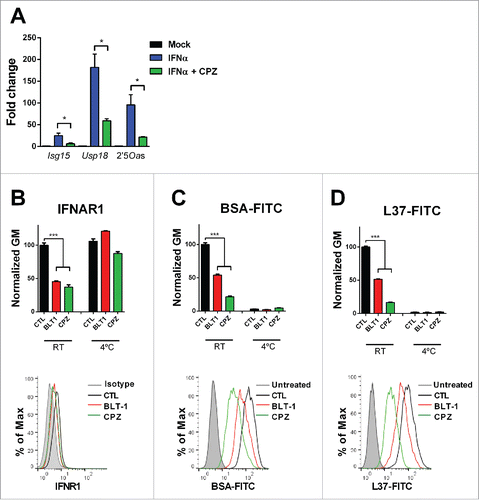
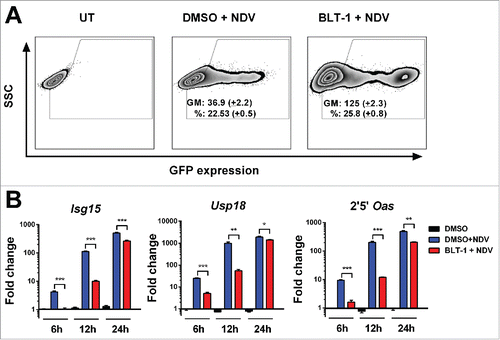
Figure 7. BLT-1 increases NDV infection. (A) Representative flow cytometry dot plot 24 h after infection with an MOI of 1 of NDV expressing GFP in L929 cells pretreated 1 h with BLT-1 (15 µM), vehicle (DMSO) or left untreated (UT). The geometric mean (GM) of GFP expression and the percentage of infection (%) are expressed as mean + SEM (number of replicates = 3). (B) Quantitative real time PCR analysis of ISGs mRNA at 6 h, 12 h and 24 h postinfection at an MOI of 1 after treatment with BLT-1 (15 µM) for 1 h. Data are expressed as mean + SEM (number of replicates = 3). (t-test. *p < 0.05, **p < 0.01, ***p < 0.001).