Figures & data
Figure 1. Increased number of cells expressing FAM13A in the tumor region of patients with NSCLC. (A) Histological arrays obtained from the control (CTR) and tumoral region (TU) of lung tissue of our NSCLC patient cohort with adenocarcinoma (ADC) and squamous cell carcinoma (SCC) subtypes as well as lung tissue array from control patients (HC). (B) Immunohistochemistry (IHC) of FAM13A on lung tissue array: control (40x CTR) and tumoral (40x, 63x TU) area of patients with ADC or SCC as well as lung tissue from control patients (40x HC). Staining with anti-FAM13A antibody is shown in brown, negative controls (63x neg CTR) of control and tumoral area are depicted in the lower panel. Diagrams show semi-quantitative analysis of the immunohistochemical staining of ADC and SCC lung section samples comparing tumoral and control area of tumor patients and control area of healthy control patients (N (ADCcontrol) = 10; N (ADCtumoral) = 10; N (SCCcontrol) = 9; N (SCCtumoral) = 9; N (HC) = 10). (C) Confocal microscopic images of lung sections after immunofluorescence of the control (63x CTR) and tumoral (63x TU) area taken from the lung of a patient with SCC and the respective negative control (63x neg. CTR). Staining with anti-FAM13A antibody is shown in red and nuclei are counterstained in blue (DAPI). Data are shown as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.
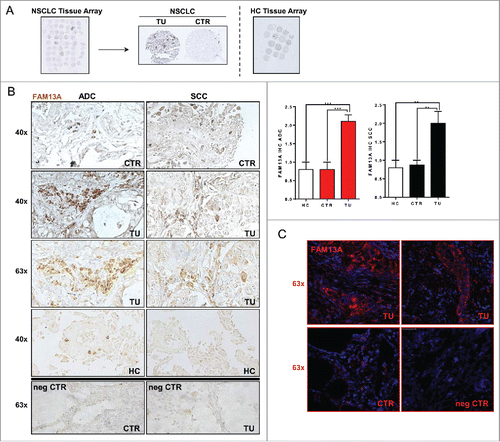
Table 1. Clinical data of the NSCLC patient cohort analyzed in this study.
Table 2. Clinical data of the cohort of control subjects analyzed in this study.
Figure 2. Increased FAM13A protein isoforms in the tumoral region of patients with NSCLC. (A) Schematic illustration of the Rho protein pathway and function of the RhoGAP domain of FAM13A. (B) Protein structure of FAM13A isoform 1 (v2) containing the RhoGAP domain (aa39-326, blue) with a total molecular weight of 117 kDa and of FAM13A isoform 2 (v1) with a molecular weight of 80 kDa. Binding sites of the anti-FAM13A antibody used in this study are depicted (aa910-971, yellow). (C) Western Blot analysis using anti-FAM13A antibody against total lung proteins isolated from the control and the tumoral area of patients with ADC and SCC (N (ADCcontrol) = 7; N (ADCtumoral) = 7; N (SCCcontrol) = 3; N (SCCtumoral) = 3). Red numbers indicate different FAM13A isoforms. (D) Western blot quantification of FAM13A total protein level, quantified by enclosing all FAM13A isoforms. Data are indicated as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.
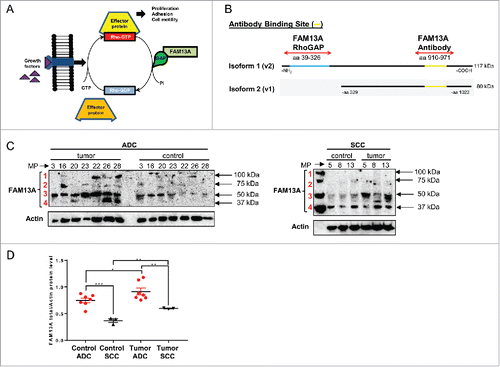
Figure 3. T regulatory cells purified from peripheral blood mononuclear cells express Foxp3 and inhibit FAM13A RhoGAP as well as HIF1α. (A) Binding sites of the primers used to analyze the expression of FAM13A isoforms 1 and 2. The first primer pair binds only the RhoGAP domain of the FAM13A isoform 1, whereas the second primer pair binds to a region that is present in both isoforms. (B) Experimental design of T regulatory cell isolation from healthy volunteers using PE labeled CD25 antibodies and anti-PE magnetic beads. (C) Purity of CD4+CD25+T cells (left panel) and CD4+CD25+Foxp3+ T cells (right panel) isolated using PE-labeled CD25 antibodies and anti-PE magnetic beads. (D) Quantitative real-time PCR analysis of FAM13ARHOGAP, TBX21, HIF1A, FOXP3, and CTLA4 in CD25− T effector cells (CD25−, N = 4) in comparison to CD25+ T regulatory cells (TREG, N = 4). (E) Correlation between CTLA4 mRNA expression and FAM13A protein level in the control region (CTR, N = 7) and tumoral region (TU, N = 9) of patients with NSCLC. (F) Correlation between FAM13A and HIF1A mRNA expression in the control region (CTR, N = 27) and tumoral region (TU, N = 24) of patients with NSCLC. Data are shown as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.
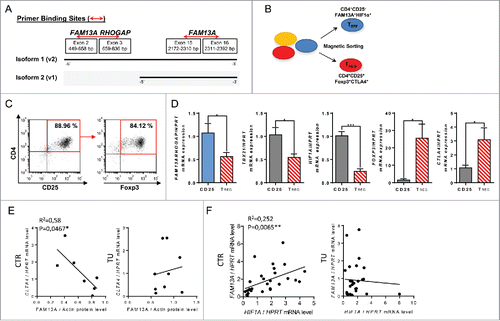
Figure 4. Treatment with TGFβ reduces FAM13A mRNA levels and induces programmed tumor cell death in A549 cells. (A) Annexin V/PI FACS analysis of untreated A549 cells (UN, N = 3) in comparison to cells treated with 3 ng/mL of TGFβ (+3 ng TGFβ, N = 3). (B) Cell count of untreated A549 cells (UN, N = 3) in comparison to A549 cells treated with 3 ng/mL TGFβ (+3 ng TGFβ, N = 3). (C–E) Quantitative real-time PCR analysis of AKT (C), HIF1A (D), FAM13A, and FAM13ARHOGAP (E) mRNA expression in untreated (N (UN) = 3) and with 3 ng/mL TGFβ (+ 3 ng TGFβ = 3) treated A549 cells. (F) Western Blot analysis using anti-FAM13A antibody in untreated and with 30 ng/mL TGFβ treated A549 cells for 24 h and 48 h (left panel). Diagrams show the quantification of FAM13A isoforms. (G) Detailed view of fluorescence staining of A549 (+ 3 ng TGFβ) cytospins (63x) using anti-FAM13A antibody (red) and DAPI (blue) to counterstain the nuclei. (H) IHC of A549 cytospins (x40) left untreated (UN) or treated with 3 ng/mL TGFβ using anti-FAM13A antibody (brown). Negative controls (neg CTR) treated with PBS are shown on the right-hand side image. (I) CFSE proliferation assay of A549 cultured for 96 h in the presence of medium (unstimulated: black lane), 3 ng/mL of TGFβ (red lane) or 30 ng/mL of TGFβ (blue lane). Data are shown as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.
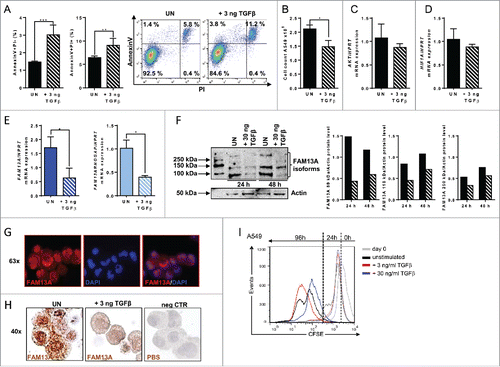
Figure 5. Treatment with TGFβ decreases A549 cell proliferation alongside an upregulation of cellular migration genes. (A, B) RNA sequencing analysis (RNA-Seq) of RNA isolated from A549 cells treated with 30 ng/mL TGFβ (+ 30 ng TGFβ, N = 3) or left untreated (UN, N = 3). The log2 fold change of the RNA expression of significantly altered genes is shown as a heatmap (A) and as a bar chart (B), (red: upregulation; blue: downregulation). Selected genes belong to different classes: tumor suppressor and apoptosis (green), cell migration (pink), tumor promoter (blue) and proliferation (yellow). (C) Ki-67 proliferation FACS analysis of untreated (UN, N = 3) or with 30 ng/mL TGFβ (+ 30 ng TGFβ, N = 3) treated A549 cells. (D) Quantitative real-time PCR analysis of FAM13A, FAM13ARHOGAP, HIF1A, and AKT mRNA expression in untreated (UN, N = 3) and with 30 ng/mL TGFβ (+ 30 ng TGFβ, N = 3) treated A549 cells. Data are shown as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.
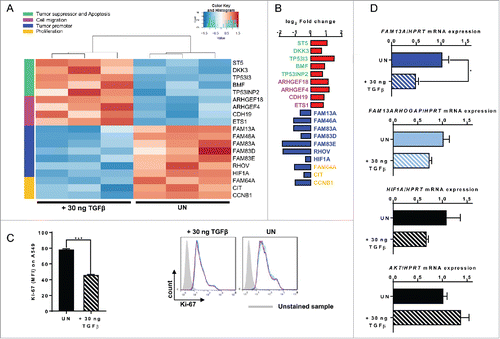
Figure 6. Targeted inhibition of FAM13A in A549 cells via siRNA. (A) Quantitative real-time PCR analysis of FAM13A, FAM13AROHGAP, and HIF1A mRNA expression in untreated A549 cells (UN, N = 3) and in A549 cells transfected with non-targeting siRNA (siNT, N = 3) or FAM13A siRNA (siFAM13A, N = 3). Lower panel: detailed view of fluorescence staining of A549 transfected with siGFP (green) and counterstained with DAPI (blue) (63x). (B, C) RNA sequencing analysis (RNA-Seq) of RNA isolated from A549 cells transfected with FAM13A siRNA (siFAM13A, N = 3) or left untreated (UN, N = 3). The log2 fold change of the RNA expression of significantly altered genes is shown as a heatmap (B) and as a bar chart (C), (red: upregulation; blue: downregulation). Selected genes belong to different classes: tumor suppressor and apoptosis (green), cell migration (pink), tumor promoter (blue) and proliferation (yellow). (D) Cell count of untreated A549 cells (UN, N = 3) and A549 cells transfected with non-targeting siRNA (siNT, N = 3) or FAM13A siRNA (siFAM13A, N = 3). (E) Ki-67 proliferation FACS analysis of untreated A549 cells (UN, N = 3), A549 cells transfected with non-targeting siRNA (siNT, N = 3) or FAM13A siRNA (siFAM13A, N = 3). (F, G) Quantitative real-time PCR analysis of TGFB (F) and TGFBRII (G) mRNA expression in untreated A549 cells (UN, N = 3) and A549 cells transfected with non-targeting siRNA (siNT, N = 3) or FAM13A siRNA (siFAM13A, N = 3). Data are shown as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.
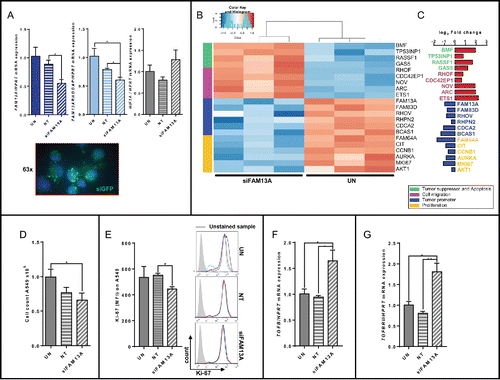
Figure 7. Increased FAM13A expression during cellular migration of A549 cells around a cell wound: role of TGFβ. (A) Scratched A549 cell layer. Photos were taken 0, 6, 12, 24, and 48 h after the scratch to detect cellular migration. Upper panels show untreated (UN, N = 3) A549 cells, lower panels A549 cells treated with 3 ng/mL TGFβ (+ 3 ng TGFβ, N = 3). The scratch is highlighted in blue. (B) Relative migration rate in percent of scratched A549 cells left untreated or treated with 3 ng/mL TGFβ. (C) Cell count of scratched A549 cells left untreated (UN, N = 3) or treated with 3 ng/mL TGFβ (+ 3 ng TGFβ, N = 3). (D–I) Quantitative real-time PCR analysis of FAM13A (D), FAM13ARHOGAP (E), HIF1A (F), AKT (G), EGFR (H), and ETS1 (I) in scratched A549 cells left untreated (UN, N = 3) or treated with 3 ng/mL TGFβ (+ 3 ng TGFβ, N = 3). Data are shown as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.
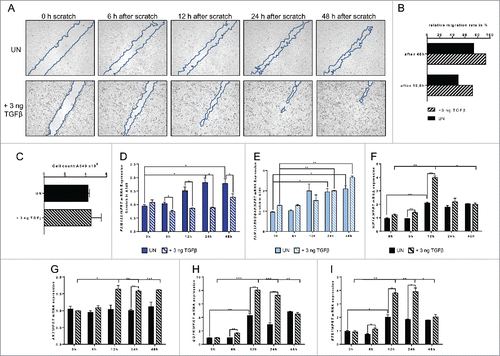
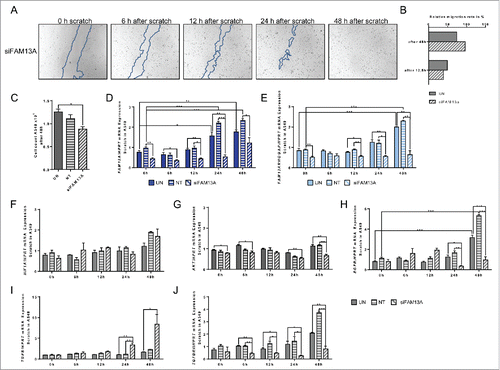
Figure 8. Effect of targeted inhibition of FAM13A via siRNA on cellular migration of A549 cells around a cell wound. (A) Scratched A549 cell layer after transfection with FAM13A siRNA (siFAM13A). Photos were taken 0, 6, 12, 24, and 48 h after the scratch to detect cellular migration. The scratch is highlighted in blue. (B) Relative migration rate in percent of scratched A549 cells left untreated (UN, N = 3) or transfected with FAM13A siRNA (siFAM13A, N = 3). (C) Cell count of scratched A549 cells left untreated (UN, N = 3) or transfected with FAM13A siRNA (siFAM13A, N = 3). (D–J) Quantitative real-time PCR analysis of FAM13A (D), FAM13ARHOGAP (E), HIF1A (F), AKT (G), EGFR (H), TGFB (I), and TGFBRII (J) in scratched A549 cells left untreated (UN, N = 3) or transfected with either NT (N = 3) or FAM13A siRNA (siFAM13A, N = 3). Data are shown as mean values ± s.e.m. using Student's two-tailed t-test *p = 0.05; **p = 0.01, ***p = 0.001.