Figures & data
Figure 1. Identification of a target RNA-NP. (A) An immortalized murine bone marrow-derived DC line (DC2.4) was transfected with GFP RNA comparing the cationic liposome DOTAP to JETPEI and JETPEI-mannose, and cells were screened one day later for assessment of GFP expression via flow cytometry. (B) C57Bl/6 mice, were given a low precursor frequency of OT-Is, and vaccinated intravenously with OVA RNA or control RNA (luciferase) encapsulated in either DOTAP, or linear polyethylenimine NPs with DC targeting mannose receptors (JETPEI-Mannose); OVA-specific T cell immunity was assessed from spleens of vaccinated mice 1 week later by flow cytometry. (C) Different ratios of GFP RNA to NP DOTAP were encapsulated and assessed for %transfection efficiency of DC2.4s in vitro. Cells were grown in vitro and harvested 1 d after addition of RNA-NPs to culture media. (D) Cationic lipid preparations of DOTAP and DOTAP-DOPE were compared with RNA-iMAX lipofectamine based on percent GFP transfection efficiency (left) and GFP-MFI (right) after 24 h. (E) Cationic lipid preparations of DOTAP and DOTAP-DOPE were compared with RNA-iMAX lipofectamine based on percent GFP transfection efficiency (left) and GFP-MFI (right) after 72 h. (F) Cationic lipid preparations of DOTAP and DOTAP-DOPE were compared with RNA-iMAX lipofectamine based on MHCI MFI 24 h after transfection of DC2.4s with GFP RNA. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, unpaired t test).
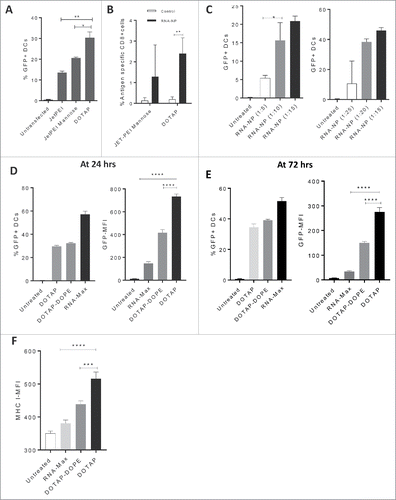
Figure 2. RNA-NP characterization, stability, and in vivo gene expression. (A) DOTAP NPs were left uncomplexed or complexed with luciferase mRNA and size was measured using cryo-electron microscopy. (B) DOTAP NPs were complexed with total RNA derived from a glioma cell line and particle size was measured using dynamic light scattering. (C) NPs encapsulating luciferase RNA were stored over time at room temperature (blue) or 37°C (red) before transfection of HEK cells. Transfection efficiency over time (relative to transfection efficiency at hour zero) is measured using bioluminescent imaging. (D) NPs encapsulating luciferase RNA were injected locally into the peritoneum of albino C57Bl/6 mice followed by peritoneal luciferin injection 6 h post-RNA-NPs. (E) NPs encapsulating luciferase RNA were injected intravenously into three C57Bl/6 mice followed by organ harvests of spleens lymph nodes, livers, kidneys, and lungs re-suspended in a 24-well plate with PBS to which luciferin substrate was added and measured for bioluminescent activity.
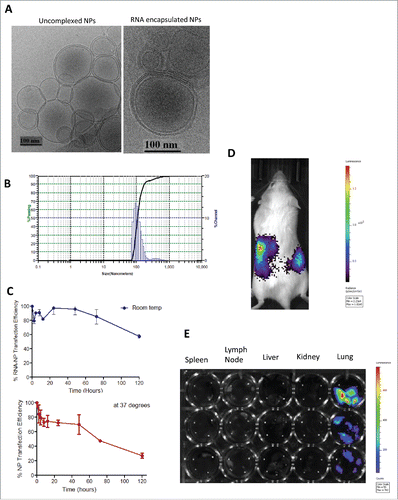
Figure 3. Antigen-specific immunogenicity of RNA-NPs. (A) C57Bl/6 mice were vaccinated with a single OVA RNA-NP complex following OT-I administration before splenocytes were harvested 1 week later. Lymphocytes were gated off by forward and side scatter, followed by gating of CD8+ T cells (APC) by side scatter. OVA-specific CD8+ cells were then identified by percent Ly5.1 positive (PercP) of CD8+ (APC) T cells (*p < 0.05; **p < 0.01, Mann–Whitney test). (B) Expansion of CFSE labeled splenocytes from OT-I transgenic mice in the presence of OVA mRNA-NPs, NPs alone, or GFP mRNA NPs. (C) C57Bl/6 mice, injected with OT-I T cells, were vaccinated with a single OVA RNA-NP complex, RNA alone or CFA-OVA peptide before splenocytes were harvested 1 week later and assessed for expansion of antigen-specific CD8+ cells (*p < 0.05; **p < 0.01, Mann–Whitney test). (D) C57Bl/6 mice were injected with OT-I T cells followed by vaccination with a single OVA RNA-NP complex, control RNA-NPs, NPs alone, RNA alone, or CFA-OVA peptide before lymph nodes were harvested 1 week later and assessed for expansion of antigen-specific CD8+ cells (*p < 0.05; **p < 0.01, unpaired t test). (E) C57Bl/6 mice were vaccinated with a single OVA RNA-NP complex following OT-I administration before splenocytes were harvested 1 week later. Splenocytes were then co-cultured with OVA peptide for 1 to 2 d, before cells were spun down for assessment of functional antigen-specific T cells by intracellular staining (right); supernatants were separated and simultaneously assessed for IFN-gamma production (left) (**p < 0.01, Mann–Whitney test).
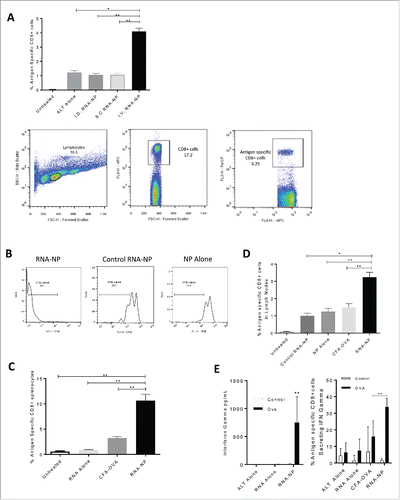
Figure 4. Systemic immune activation of RNA-NPs. (A) NPs were complexed with GFP RNA and injected i.v. into C57Bl/6 mice. Spleens were harvested 1 d after injection for assessment of CD11c+ cells expressing GFP by flow cytometry (*p < 0.05; **p < 0.01, Mann–Whitney test). (B) NPs alone or RNA-NPs were administered to C57Bl/6 mice bearing subcutaneous B16F10 melanomas and serum was collected 6 h later for assessment of IFN-α levels by ELISA (upper limit of detection for assay is (2,000 pg/dL) (*p < 0.05, Mann–Whitney test) (C) RNA-NPs were administered systemically to C57Bl/6 mice and spleens were collected within 24 h for assessment of MHC I (****p < 0.0001, unpaired t test) and MHC II (**p < 0.01, Mann–Whitney test) on CD11c+ splenocytes. (D) RNA-NPs were administered systemically to C57Bl/6 mice and spleens were collected within 24 h for assessment of CD80, CD86, and CD40 on CD11c+ splenocytes (*p < 0.05; **p < 0.01, Mann–Whitney test). (E) RNA-NPs were administered systemically to C57Bl/6 mice and livers were collected within 24 h for assessment of CD80 and CD86 on CD11c+ liver WBCs (*p < 0.05; **p < 0.01, Mann–Whitney test). (F) Intravenous RNA-NPs were administered to C57Bl/6 mice and lymph nodes and bone marrow were harvested within 24 h for assessment of %CD86 on CD11c+ cells (*p < 0.05; **p < 0.01, unpaired t test). (G) Intravenous RNA-NPs were administered to C57Bl/6 mice 2 weeks after subcutaneous implantation of B16F10 (1 × 106 /mouse); tumors were harvested within 24 h for assessment of %CD86 on CD11c+ cells (*p < 0.05, unpaired t test).
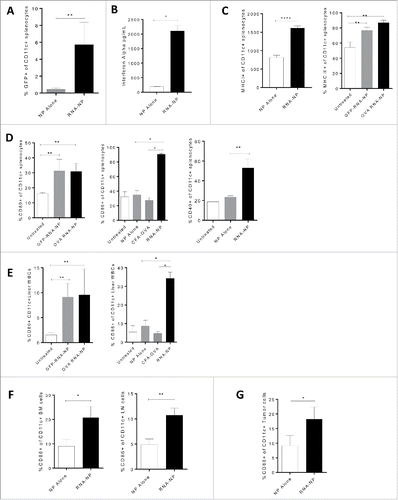
Figure 5. Antitumor efficacy from RNA-NPs and capacity for immunomodulatory modification. (A) OVA RNA-NP complexes were injected once weekly (× 3) into naïve C57Bl/6 mice starting 5 d after intracranial implantation of B16F10-OVA (6,250 cells/mouse) (**p < 0.01, Gehan–Breslow–Wilcoxon test). (B) DC2.4 cells were grown in culture and transfected with GM-CSF RNA-NPs. One day later, cells were spun down and supernatants were collected for analysis of GM-CSF production by ELISA (*p < 0.05, unpaired t test). (B) C57Bl/6 mice were injected with OT-I T cells followed by a single RNA-NP complex co-encapsulated with or without GM-CSF RNA and spleens were harvested 1 week later for analysis of %antigen-specific CD8+ cells (*p < 0.05; **p < 0.01, Mann–Whitney test). (C) C57Bl/6 mice were injected with OT-I T cells followed by vaccination with a single RNA-NP (GFP RNA-NP, OVA RNA-NP, or OVA+ GM-CSF RNA-NP) complex 5 d after intra-cranial B16F10-OVA (6,250 cells/mouse) implantation (*p < 0.05; **p < 0.01, Gehan–Breslow–Wilcoxon test). (D) C57Bl/6 mice were injected with OT-Is followed by vaccination with a single RNA-NP (GFP RNA-NP, OVA RNA-NP, or OVA+ GM-CSF RNA-NP) complex 1 d after subcutaneous B16F10-OVA tumor (1 × 106 /mouse) implantation (*p < 0.05; **p < 0.01, Mann–Whitney test).
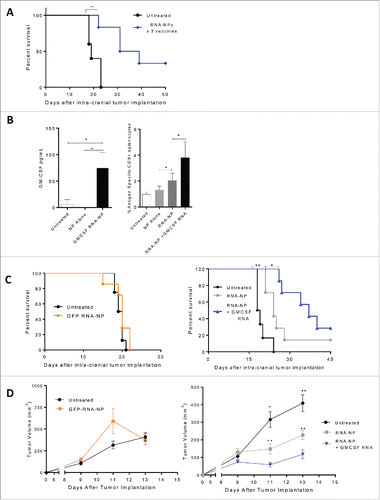
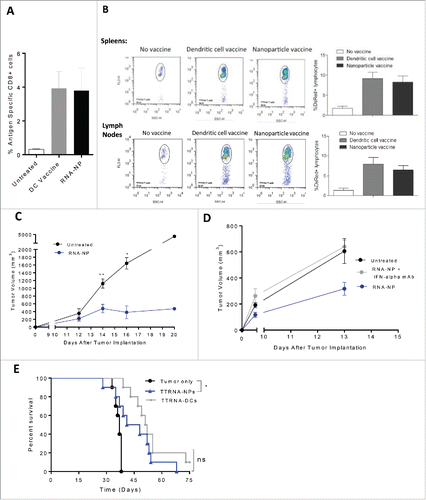
Figure 6. Efficacy of RNA-NPs targeting physiologically relevant antigens. (A) C57Bl/6 mice were vaccinated with a single OVA RNA-NP or DC vaccine pulsed with OVA mRNA following OT-I administration before splenocytes were harvested 1 week later for assessment of %antigen-specific CD8+ cells. (B) DS red+ tumor-specific T cells were expanded in vitro from primed spleens of mice immunized against TTRNA extracted from KR158B-luc. These T cells were adoptively transferred after 5–7 d of in vitro activation into KR158B-luc intracranial tumor-bearing C57Bl/6 mice followed by TTRNA-NPs or TTRNA-pulsed DCs. (C) C57Bl/6 mice were implanted with B16F0 melanomas (250,000 cells/mouse) in the flank and vaccinated 1 d later with weekly TTRNA-NPs × 3 (*p < 0.05; **p < 0.01, Mann–Whitney test). (D) C57Bl/6 mice were implanted with B16F0 melanomas (250,000 cells/mouse) in the flank and vaccinated 1 d later with weekly TTRNA-NPs that were pre-treated with an IFN-α-blocking antibody. (E) C57Bl/6 mice were stereotactically implanted with KR158B-luc astrocytoma cells and received a single dose of 9Gy TBI (on Day 4) followed by an i.v. injection of 5 × 104 lineage negative bone marrow-derived stem cells (within 6 h of TBI) and i.v. injection of 107 tumor-specific T lymphocytes (1 d post-TBI).Citation17 This was immediately followed by vaccination of 2.5 × 105 total tumor RNA-pulsed DCs or TTRNA-NPs. The second and third RNA-NP/DC vaccines were administered at weekly intervals (*p < 0.05, Gehan–Breslow–Wilcoxon test).