Figures & data
Figure 1. Diminished NK cell effector functions in T-ALL patients. PBMC from patients or healthy controls were co-incubated with K562 target cells in the presence of anti-CD107a mAb for 6 h. NK cells were gated as CD14/CD19−CD56dimCD3− cells, and assessed for (A) degranulation by measuring percentage CD107a+ NK cells, or (B) percent intracellular IFNγ expression (n = 3 T-ALL, n = 13 controls, ± SEM). Statistical significance was calculated using the non-parametrical Mann–Whitney test. (C) Frequencies of CD8+ CD3−CD56dim peripheral blood NK cells from healthy children or adult controls compared with pediatric or adult T-ALL patients. Data are presented as percentages ± SEM. Statistical significance was calculated using the non-parametrical Mann–Whitney test.
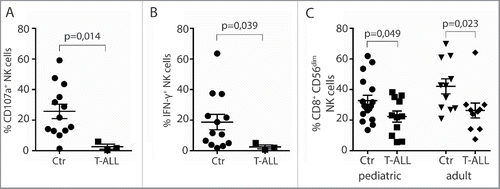
Figure 2. Reduced frequencies of NK cells with activating receptors in T-ALL patients. (A) Frequencies of CD56dimCD3− NK cells expressing NKG2D, NKp46, DNAM-1, NKG2A, and NKR-P1A were analyzed on CD3−CD56dim peripheral blood NK cells from healthy children or adults, and pediatric or adult T-ALL patients. Data are presented as percentages ± SEM. (B) Relative fluorescence index (RFI) of NKG2D, NKp46, DNAM-1, NKG2A, and NKR-P1A were calculated by dividing the median fluorescence intensity of each receptor on CD3−CD56dim peripheral blood NK cells to negatively stained populations with the same antibody. Data are presented as RFI values ± SEM. Statistical significance was calculated using the non-parametrical Mann–Whitney test.
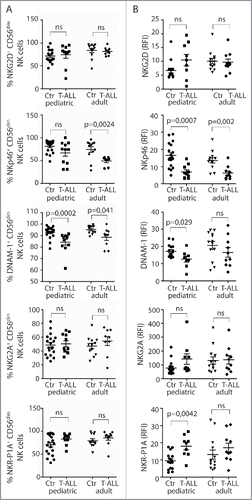
Figure 3. Low NK-cell responses and skewed receptor repertoires in rats with RL. (A) Flow cytometric analysis of the distribution of Ly49s3+, NKR-P1Bdim, or NKR-P1Bbright NK cells in blood, spleen, and bone marrow isolated from control rats (n = 9) or rats with RL (n = 10). Data represent the average of six independent experiments ± SEM. (B) Degranulation of NK cells from healthy rats (n = 6), rats with blast load <2% of PBMC (n = 3), or >30% of PBMC (n = 4) in response to YAC-1. NK cells were gated as NKR-P1A+CD3− cells. Data represent the average of three independent experiments ± SEM. Intracellular IFNγ production by NKR-P1A+CD3− NK cells was analyzed by flow cytometry in samples stimulated for 6 h by (C) the indicated plate-bound antibodies or (D) IL-2 alone or in combination with IL-12 or IL-18 using healthy control rats (n = 6), rats with blast load <2% of PBMC (n = 3), rats with blast load >30% of PBMC (n = 3). Values represent the average of three independent experiments ± SEM. MFI analysis of (E) NKG2D or (F) NKp46 expression on NKR-P1A+CD3− NK cells from control rats (n = 4) or rats with RL (n = 5). Values represent the average of three independent experiments. (G) qRT-PCR analysis of RL (n = 4), primary T cells (n = 4), and YB2/0 cells (n = 4). Statistical significance was calculated using the non-parametrical Mann–Whitney test.
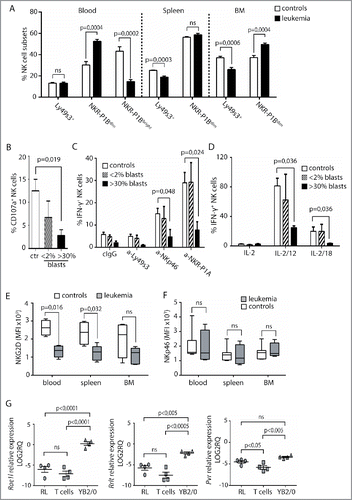
Figure 4. Cytokine pre-activation upregulates activating receptors and promotes NK cell lysis of RL. (A) Degranulation of primary enriched splenic NK cells in response to YAC-1 or RL. Values represent data from three independent experiments (n = 9) ± SEM. (B) Specific lysis of YAC-1 or RL target cells measured by 4 or 20 h 51Cr release assay using enriched, splenic NK cells. Values represent the average of triplicates of one representative experiment out of four independent experiments. (C) qRT-PCR analysis of Dnam1 expression in primary NK cells (n = 4), NK cells pre-cultured overnight in medium with IL-15 (n = 3), or in medium with IL-15, IL12, and IL-18 (n = 3). Statistical significance was calculated using the Kruskal–Wallis test. (D) MFI analysis of CD25, NKG2D, Ly49s3, FasL, and NKp46 on NK cells after overnight culture in medium alone (n = 3), with IL-15 alone (n = 3), or with IL-15, IL12, and IL-18 (n = 3). Data represent the average of three independent experiments ± SEM. Statistical significance was calculated using the non-parametrical Mann–Whitney test. (E) Specific lysis of YB2/0 or RL measured by 51Cr release assay with enriched splenic NK cells pre-cultured overnight in medium alone, with IL-15 alone, or with IL-15, IL12, and IL-18. NKG2D antibody or isotype control was pre-incubated with NK cells 30 min before assay. Data represent average of triplicates of one representative experiment of three independent experiments.
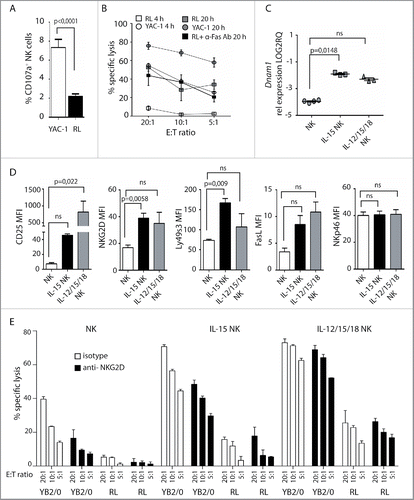
Figure 5. Adoptive transfer of cytokine pre-activated NK cells reduces RL load. Rats (n = 10) were sub-lethally irradiated at 4 Gy and injected with RL 24 h later. IL-12, IL-15, and IL-18 pre-activated NK cells were adoptively transferred to rats (n = 5) at days 3, 6, and 9. Control rats received no NK cells (n = 5). (A) Spleen weights of rats having RL with (white bars, n = 5) or without (black bars, n = 5) infusion of pre-activated NK cells. (B) Total numbers of RL per mL blood or total numbers of RL per spleen in rats with (white bars, n = 5) or without (black bars, n = 5) infusion of pre-activated NK cells. (C) Percent reduction of RL blasts in spleen, blood, or BM in rats receiving pre-activated NK cells. (D) Donor NK cells (CD45.2neg) are readily detected in spleen, blood, and BM at sacrifice 4 weeks after RL injection. (E) Total numbers of donor CD45.2neg NK cells per spleen or per mL blood at sacrifice 4 weeks after RL injection. (F) Percent proliferating Ki67+ host or donor NK cells at sacrifice 4 weeks after RL injection. (G) Expression of FasL, NKG2D, Ly49s3, and NKp46 on host and donor NK cells at sacrifice 4 weeks after RL injection, presented as RFI values ± SEM. Statistical significance was calculated using the non-parametrical Mann–Whitney test. Data represents two individual experiments with a total five rats in each group.
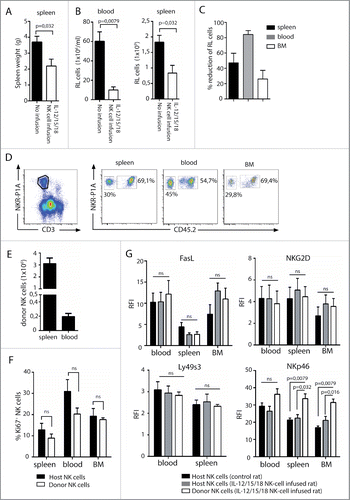
Table 1. List of primers.
