Figures & data
Figure 1. Vaccination of RENCA-bearing mice against DLK1 results in slowed tumor growth, the loss of DLK1+ pericytes in the TME and a compensatory increase in DLK2 expression by tumor-associated vascular pericytes. (A) Female Balb/c mice with established day 10 s.c. RENCA tumors on their right flanks were treated with s.c. injection (left flank) of PBS, 106 DCs transduced with rAd.IL12 (i.e., DC.IL12) or DC.IL12 pulsed with DLK1-derived peptide epitopes (i.e., DC.IL12.DLK1) per Materials and methods. An identical s.c. booster vaccination was provided on day 17 post-tumor inoculation. (B) Tumor growth was monitored every 3–4 d and is reported as the mean ± SEM for five animals per group. *p < 0.05, two-way ANOVA. (C) On day 21, tumor tissues and (tumor-uninvolved kidneys from matched animals) were harvested and then digested mechanically and enzymatically as described in the Materials and methods, yielding single-cell suspensions. Live cells (DAPI negative) cells were flow-sorted to select for CD45−CD146+CD34− pericytes and CD45−CD146+CD34+ VEC populations. (D) Total mRNA was isolated from sorted populations of pericytes and VECs and analyzed for DLK2 expression by quantitative real-time PCR. Relative mRNA expression was normalized to HPRT1 expression. *p < 0.05, one-way ANOVA compared with kidney pericyte DLK2 levels. In (E), RENCA tissue sections were analyzed for expression of DLK2 (green) in NG2+ pericytes (red) by immunofluorescence microscopy. Scale bar = 1mm. All data are representative of those obtained in two independent experiments.
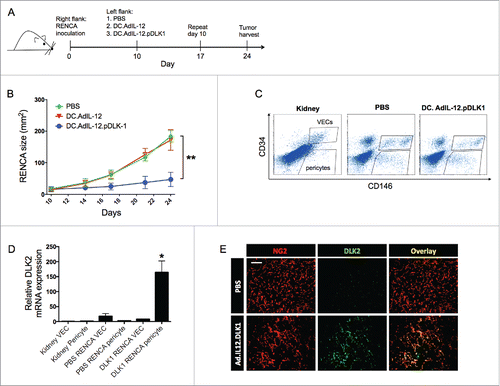
Figure 2. Coordinate vaccination with lvDLK1 and lvDLK2 is immunogenic and therapeutic in the RENCA model. Female Balb/c mice bearing established day 7 s.c. RENCA tumors were treated with i.d. injection of PBS, 104 lvNEG, 104 lvDLK1, 104 lvDLK2 or 104 lvDLK1 + 104 lvDLK2. (A) Tumor growth was monitored every 3–4 d through day 20, at which time the animals were killed. Tumor sizes are reported as the mean ± SEM for five animals per group. *p < 0.05, two-way ANOVA. Tumors were then harvested to generate tumor sections or they were mechanically/enzymatically-digested to generate single-cell suspensions. (B) Tumor sections were analyzed by immunofluorescence microscopy for NG2+ pericytes (red) and infiltrating CD8+ T cells (green). Using Metamorph software (Materials and methods), CD8+ T cell infiltrates and NG2+ pericytes in the images were quantified and reported as mean integrated fluorescence ± SEM. *p < 0.05 and **p < 0.01, one-way ANOVA (C) Flow cytometry was also performed on the resulting cell suspensions to quantify Tbet+ CD8+ TIL. Tumor sections were analyzed for CD8+ TIL (green) infiltration and NG2+ (red) expression. In (D), splenocytes harvested from day 21 tumor-beating mice were stimulated in vitro with syngeneic DC infected with rAd.DLK1 (DC.DLK1), rAd.DLK2 (DC.DLK2) or a mixture of both DC populations (i.e., DC.DLK1 + DC.DLK2) for 5 d. Responder T cells were the isolated and restimulated with control or rAd-infected DC as indicated for 18h, and the resultant culture supernatants analyzed for IFNγ content using a cytokine-specific ELISPOT assay. *p < 0.05, one-way ANOVA. In (E) and (F), tumor sections were analyzed for NG2+ pericyte co-expression of DLK1 and DLK2, respectively, via immunofluorescence microscopy. Scale bar = 1mm. Data in A, B, C, E and F are representative of those obtained in three independent experiments. Data in D is representative of two independent experiments.
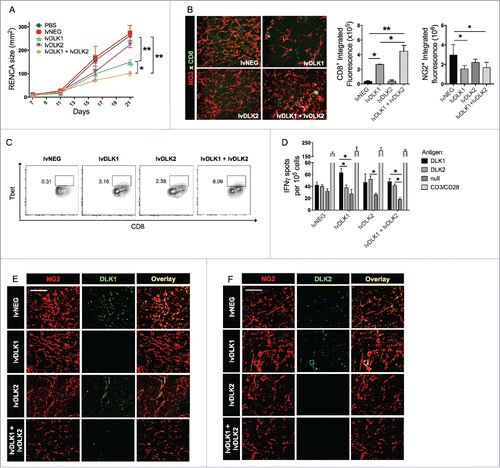
Figure 3. Coordinate vaccination with lvDLK1 and lvDLK2 results in tumor VN. (A) RENCA tumor sections from were stained for NG2+ pericytes (red) and CD31+ VECs (green) to characterize blood vessel morphology. (B) Mean vessel density was determined by quantitating distinct (green) VECs or VEC clusters. (C) Colocalization of NG2+ (red) pericytes and CD31+ (green) VECs was determined using Metamorph software. (D–E) RNA extracted from Day 21 RENCA tumors was analyzed for HIF2α, VEGFR1, VEGFR2 and FGFR1 transcript levels by quantitative real-time PCR. In (E), tumor CD31+ VEC (green) were analyzed for co-expression of VCAM1 (red) by immunofluorescence microscopy. Scale bar = 1mm. *p < 0.05 and **p < 0.01, one-way ANOVA compared with the negative control (lvNEG). All data are representative of those obtained in three independent experiments.
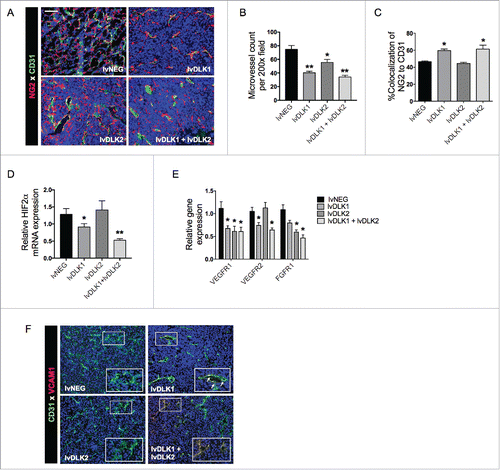
Figure 4. Coordinate vaccination with lvDLK1 and lvDLK2 results in a superior level of reduction in Treg and MDSC content in the therapeutic TME. Day 21 tumors from untreated or treated mice per were dissociated into single-cell suspensions and analyzed by flow cytometry for (A) CD11b+Gr1+ MDSC and (B) FoxP3+CD4+ Treg populations. Data are representative of those obtained in three independent experiments.
Figure 5. PD-L1 blockade fails as a monotherapy against RENCA tumors, but improves the antitumor efficacy of combined lvDLK1 + lvDLK2 vaccination. (A) CD8+ TILs and (B) bulk RENCA single-cell suspensions (per ) were analyzed for expression of PD-1 and PD-L1, respectively, by flow cytometry. In (C), mice bearing established day 7 RENCA tumors (right flank) were vaccinated i.d. with lvDLK1 + lvDLK2 (left flank) as a therapeutic vaccine. Anti-mPD-L1 blocking antibody was injected i.p. on days 7, 10, 13 and 17 post-tumor inoculations as described in Materials and methods. Tumor growth was monitored longitudinally and is reported as the mean ± SEM for five animals per group. In (D), RENCA tumors were harvested on day 26 and dissociated single-cell suspensions analyzed for (D) Tbet+CD8+ TILs and (E) FoxP3+CD4+ Tregs by flow cytometry. *p < 0.05 and **p < 0.01.
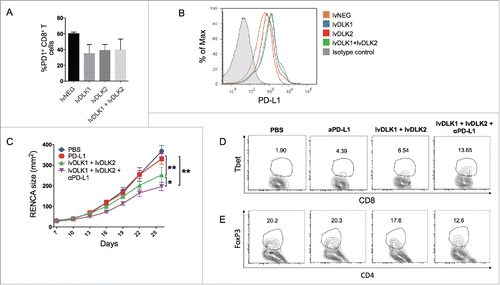
Table 1. Murine qPCR primers.
