Figures & data
Table 1. Clinicopathological characteristics at time of surgery for patients in the proteomic study.
Figure 1. Bar diagram showing the percentage of TNBC tissue samples exhibiting high amount (IHC-score 3), intermediate (IHC-score 2), low (IHC-score 1), no (IHC-score 0) CD8+ TILs in the entire TNBC cohort (TMA cohort) and the discovery mass spectrometry TNBC cohort (Cohort 1).
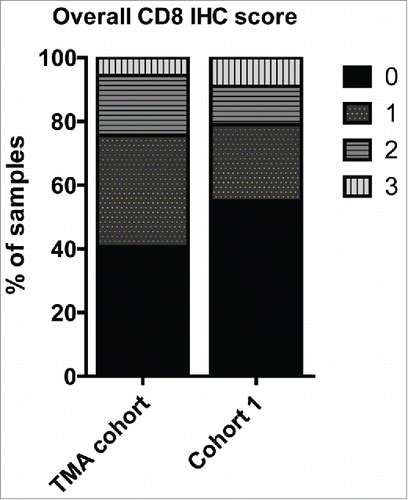
Figure 2. Hierarchical cluster of proteins differentially expressed (p < 0.05) between recurrence and recurrence-free patients with an FDR < 1% identified by Student's t-test. Red = high expression in recurrence-free vs. recurrence patients; Green = low expression in recurrence-free vs. recurrence patients.
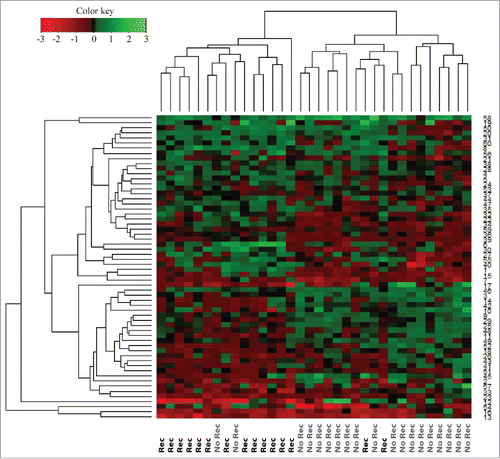
Figure 3. The proteins exhibiting significantly altered average protein expression (p ≤ 0.05) in the recurrence and recurrence-free group was examined by comparative analysis using IPA (Ingenuity Pathway Analysis). Diseases and Bio Functions revealed activation z-scores for patients with recurrence in “migration of cells,” “invasion of cells” and “proliferation of cells” compared with recurrence-free patients. Recurrence-free patients exhibited higher activation of “apoptosis” and “cell death” compared with patients with recurrence.
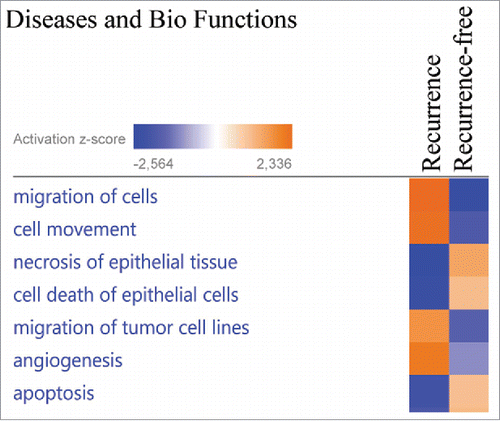
Figure 4. Network analysis obtained from Ingenuity Pathway Analysis (IPA) of aberrantly (p ≤ 0.05) regulated proteins between recurrence (R) and recurrence-free (RF) patients. Official gene symbols and relative expression values for each protein are depicted. For detailed information on aberrantly regulated proteins see Table S3.
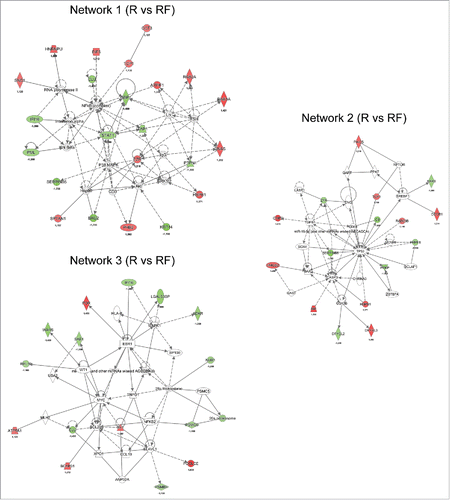
Figure 5. Network analysis obtained from IPA of differentially regulated proteins (p ≤ 0.05) between recurrence (R) and recurrence-free (RF) patients. Official gene symbols and relative expression values (recurrence-free vs. recurrence) for each protein are depicted.
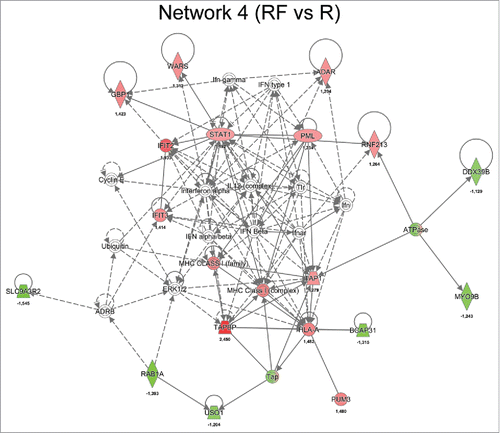
Figure 6. Kaplan-Meier survival curves showing strong association between expression of selected gene and recurrence-free survival (RFS) of TNBC patients. (A) TAP1, (B) STAT1, (C) WARS, (D) GBP1 and (E) RAD23B. Plots were generated using KMplotterCitation68 with optimal cutoff values and 10-y follow-up period.
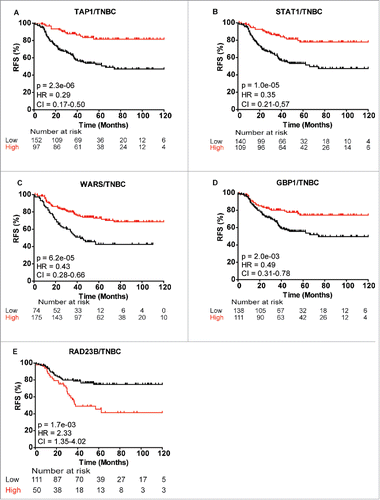
Figure 7. Kaplan-Meier curves showing association between TAP1 gene expression and recurrence-free survival (RFS) of different patient subgroups. Strong association between TAP1 expression and RFS in (A) basal-like breast cancer patients, TNBC patients with (B) lymph node-positive (N+) and (C) lymph node-negative (N−) status, respectively, and basal-like breast cancer patients with (D) N+ and (E) N− status, respectively. (F) Gene expression analysis of 10 patients from the MS data set revealed a good correlation between mRNA and protein expression of TAP1. Kaplan-Meier curves were generated with KMplotter using optimized cutoff values (A–E) and censuring at 10 y or median cutoff value (F).
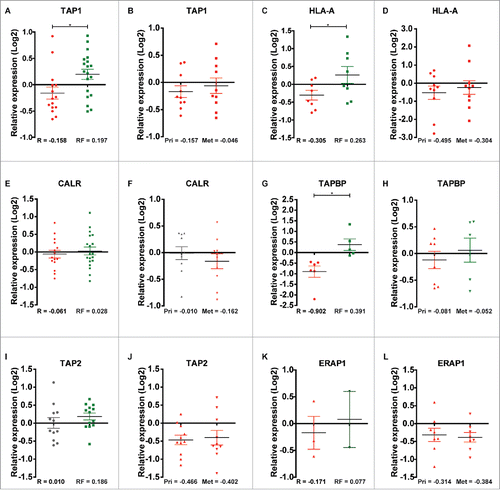
Figure 8. Relative quantile normalized expression ratios for MHC class-I-associated proteins in patient samples from the discovery and second cohorts. TAP1 shows differential regulation between (A) recurrence and recurrence-free patients and overall downregulation in (B) Pri and Met samples. HLA-A displays the same tendency as TAP1 in (C) recurrence and recurrence-free patients and in (D) Pri and Met samples. CALR shows a slight differential regulation between (E) recurrence and recurrence-free samples and a neutral to downregulated profile for (F) Pri and Met, respectively. TAPBP shows differential regulation between (G) recurrence and recurrence-free samples and differential regulation between (H) Pri and Met groups. TAP2 displayed a neutral and an overall upregulated expression status in (I) recurrence and recurrence-free patients respectively and a clear downregulation in (J) Pri and Met patient samples. ERAP1 display a tendency of differential regulation in (K) recurrence and recurrence-free samples (n = 3 and 2, respectively) and is highly downregulated in (L) Pri and Met samples. *p < 0.05.