Figures & data
Figure 1. CXCR3 ligand mRNAs are sensitive indicators of specific immune responses. (A) Peripheral blood T cells from a healthy donor were expanded in vitro and retrovirally transduced with the NY-ESO-1p157–165/HLA-A0201-specific TCR. The CD8+ T cells positively stained by the NY-ESO-1 p157–165/A0201 tetramer were isolated by sorting. (B) Following overnight incubation of NY-ESO-1p157–165 specific CD8+ T cells with 397mel or 397melA0201, the levels of 48 cytokines/chemokines in the culture supernatants were evaluated using the Bio-Plex system. Data for nine selected cytokines/chemokines that increased in an HLA-A2 dependent manner are shown. (C) PBMC from A24+ donors latently infected with EB virus were stimulated with EBNA3A246–254 (RYSIFFDYM) peptide or DMSO as a control, and total RNA was extracted at the indicated time points. The fold increase of mRNA levels of the nine selected cytokines/chemokines and of CXCL11 compared with the DMSO control was evaluated by RT-qPCR. Expression of each gene was normalized to that of GAPDH. One representative data set out of three independent experiments is shown. Data represent relative quantity means ± SD.
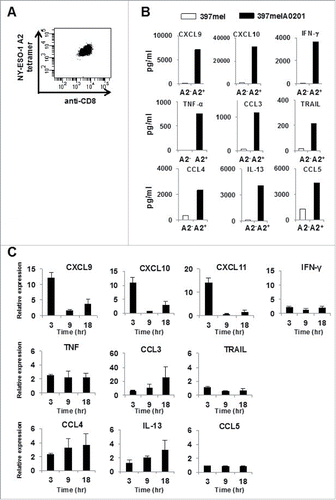
Figure 2. Kinetics of CXCR3 ligand mRNA synthesis following treatment with long peptides. (A) PBMC from A24+ donors latently infected with EB virus were stimulated with EBNA3A240–259 (20-mer; VQSCNPRYSIFFDYMAIHRS) peptide or DMSO as a control. Relative fold increase of mRNA levels of CXCL9 and IFNγ compared with the DMSO control was evaluated by RT-qPCR. One representative data out of three independent experiments is shown. Data represent relative quantity means ± SD. (B) In this experiment, we used the same PBMC aliquot from which we had previously established an HLA-B35 restricted NY-ESO-1p94–104 peptide specific CTL clone. After addition of NY-ESO-1p91–110 (20-mer) peptide or DMSO as a control, total RNA was extracted from PBMC at the indicated time points. The relative fold increase of mRNA levels of CXCL9 and IFNγ compared with DMSO control was quantified.
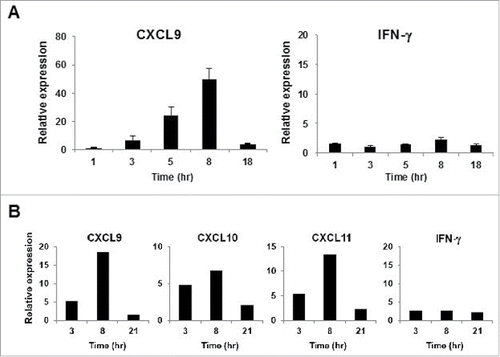
Figure 3. Kinetics of CXCR3 ligand mRNA synthesis are similar in murine and human immune system. (A) Splenocytes from naive BALB/c mice were mixed with splenocytes isolated from DUC18 mice. Mixed splenocytes were incubated with mERK2–9m peptide, wERK2–9m peptide or DMSO as a control. The relative fold increase of mRNA levels of murine CXCL9 compared with the DMSO control was quantified by RT-qPCR at the indicated time points. (B) CHP-NY-ESO-1 was subcutaneously injected into the back of BALB/c mice, twice at a 1-week interval. Splenic cells from pooled spleens (n = 3 per experiment) were prepared for analysis 7 d after the second immunization. Pooled splenic cells from naive BALB/c (left) or immunized BALB/c (right) mice were incubated with one of 17 overlapping peptides spanning the whole amino acid sequence of NY-ESO-1 (Table S1) or DMSO as a control for 8 h. Total RNA was extracted, and the relative fold increase of mRNA levels of murine CXCL9 compared with DMSO was quantified.
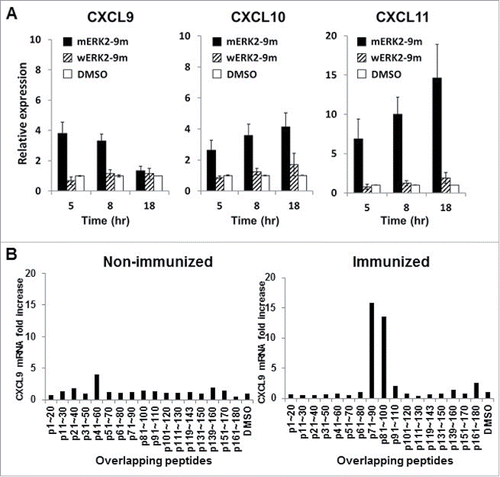
Figure 4. A minute amount of IFNγ secreted by CD8+ T cells is sufficient to induce rapid and robust CXCR3 ligand mRNA synthesis. (A) PBMC from A24+ donors latently infected with EB virus were incubated with EBNA3A246–254 peptide. At the indicated time points, the levels of CXCL9, CXCL10 and IFNγ in the culture supernatant were evaluated by ELISA. One representative data out of three independent experiments using different PBMC donors is shown. Data represent means ± SD. (B) Blocking assay, whereby PBMC from A24+ donors latently infected with EB virus were cultured for 1 h in the presence of an anti-IFNγ receptor mAb, an anti-IFNγ mAb or an isotype control mAb before incubation with EBNA3A246–254 peptide or DMSO as a control. After 5-h incubation with EBNA3A246–254 peptide, the relative fold increase of mRNA levels of CXCL9 compared with DMSO was quantified. Data represent relative quantity means ± SD. Asterisks indicate statistically significant differences (p < 0.01). (C) Flow cytometry analysis before (left) and after (right) cell sorting. The CD8+ population was depleted from PBMC by cell sorting. (D) Each population was incubated with EBNA3A246–254 peptide. The relative fold increase in mRNA levels of CXCL9 over the DMSO control was quantified. Data represent relative quantity means ± SD. Asterisks indicate statistically significant difference (p < 0.01).
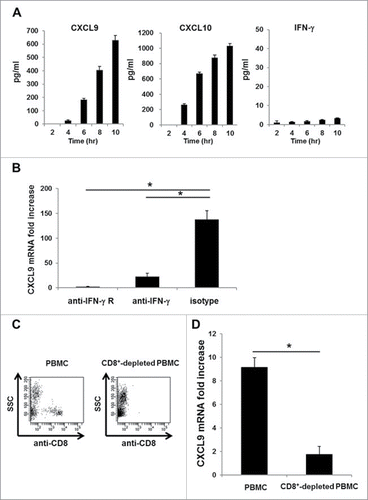
Figure 5. Detection of cellular immune responses against tumors in tumor-bearing mice. (A) Splenocytes from CT26 tumor-bearing BALB/c mice were incubated with mERK2–9m peptide, AH1 peptide, or DMSO as a control. The relative fold increase in mRNA levels of murine CXCL9 compared with DMSO was quantified 5 h later. One representative data out of three independent experiments is shown. Data represent relative quantity means ± SD. Asterisks indicate statistically significant differences (p < 0.01). (B) After subcutaneous inoculation with 1 × 106 CMS5 cells, mice were randomly divided into 2 groups. One group was left untreated, and the other was intraperitoneally administered with 100 μg of anti-CTLA-4, 200 μg of anti-PD-1 and 150 μg of anti-GITR mAbs at days 7, 9, and 11. Both mouse groups were sacrificed at day 21 and splenic cell suspensions were prepared from pooled spleens (n = 3 per experiment). Splenic cells (1 × 106) from the antibody-treated group were incubated with mERK2–9m peptide, AH1 peptide, or DMSO as a control. The relative fold increase in mRNA levels of murine CXCL9 compared with DMSO was quantified 5 h later. One representative data set out of three independent experiments is shown. Data represent relative quantity means ± SD. Asterisks indicate statistically significant differences (p < 0.01). (C) For IFNγ intracellular staining, splenic cells from both mouse groups were incubated with mERK2–9m peptide, AH1 peptide or DMSO as a control for 15 min at room temperature, and subsequently with GolgiPlug for 4 h. Following stimulation, cells were stained for cell surface CD8+ and intracellular IFNγ. Representative dot plots gated on CD8+ splenocytes were shown. These experiments were repeated three times with similar results. (D) Pooled splenic cells from antibody treated or untreated CMS5 tumor-bearing mice were subjected to IFNγ ELISPOT assays. Pooled splenic cells were incubated in vitro with mERK2–9m, AH-1 or DMSO for 18 h. One representative data out of three independent experiments is shown.
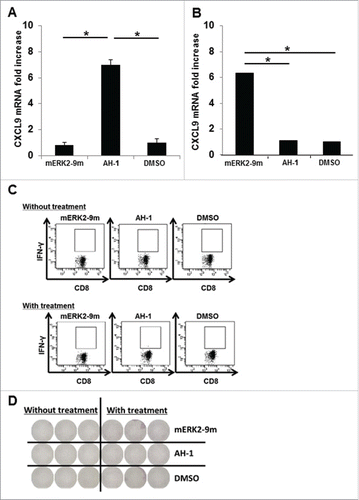
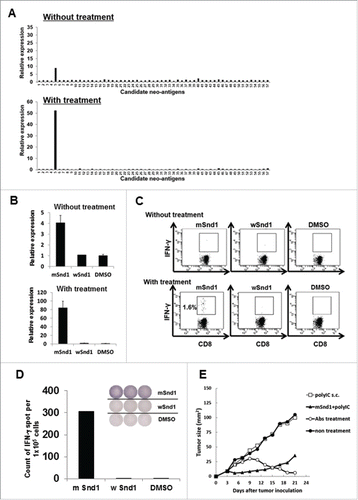
Table 1. The list of candidate neo-epitopes encoded by CMS7 tumor.
Figure 6. Successful identification of an immunogenic neo-epitope encoded by mouse sarcoma CMS7. (A) After subcutaneous inoculation with CMS7 cells, mice were randomly divided into two groups. One group was left untreated, and the other was administered intraperitoneally with anti-CTLA-4, anti-PD-1 and anti-GITR mAbs on days 7, 9, and 11. Both mouse groups were sacrificed at day 21 and splenic cell suspensions were prepared from pooled spleens (n = 3 per experiment). Splenic cells from untreated (upper) or treated mice (lower) were incubated with each panel of neo-epitope peptides for 5 h and the fold increase in CXCL9 mRNA levels compared with DMSO was quantified. One representative data set out of three independent experiments is shown. (B) Splenic cells from untreated (upper) or treated mice (lower) were incubated with mutated Snd1 peptide, its wild-counterpart or DMSO as a control for 5 h and the fold increase in CXCL9 mRNA levels compared with DMSO was quantified. (C) Splenic cells from untreated (upper) or treated mice (lower) were incubated with mutated Snd1 peptide, its wild-counterpart or DMSO as a control for 15 min at room temperature and subsequently with GolgiPlug for 4 h. Following stimulation, cells were stained for cell surface CD8+ and intracellular IFNγ. Representative dot plots gated on CD8+ splenic T cells are shown. The number indicates the percentage of CD8+ IFNγ+ T cells. These experiments were repeated three times with similar results. (D) CD8+ splenic T cells were obtained from antibody-treated mice pooled spleens by positive enrichment using the MACS system, and were stimulated in vitro with mutated Snd1-pulsed CD8− splenic cells. Cultured CD8+ splenic T cells were subjected to ELISPOT assays 10 d later. The target cells were CD8− splenic cells pulsed with mutated Snd1 peptide, or its wild counterpart. Splenic cells pulsed with DMSO were used as control targets. (E) Age-matched female BALB/c mice were inoculated with CMS7 at day 0 following two injections (at days −14 and −7) with mSnd1 peptide and poly (I:C) formulated in PBS or poly (I:C) alone using prophylactic schedules. The tumor size was monitored three times a week. Each group consisted of five mice. Mice without any immunization and mice treated by the co-administration of antibodies (anti-CTLA-4/PD-1/GITR Abs) at days 7, 9, 11 served as a negative control group and a positive control group, respectively.