Figures & data
Table 1. Clinical characteristics of the MD Anderson Cancer Center CTCL patients in the study.
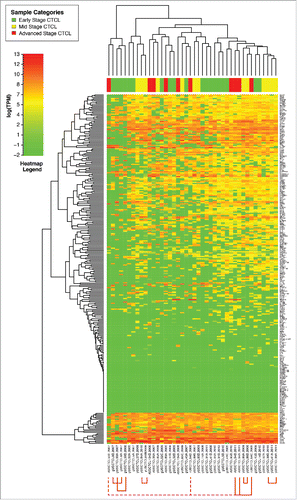
Figure 1. Unsupervised clustering analysis based on TruSeq targeted RNA gene expression of 284 select genes in skin tag lesions/benign inflammatory dermatoses (green), freshly-obtained and liquid nitrogen snap-frozen CTCL samples (gray) and FFPE CTCL tissues (yellow, light red and dark red).
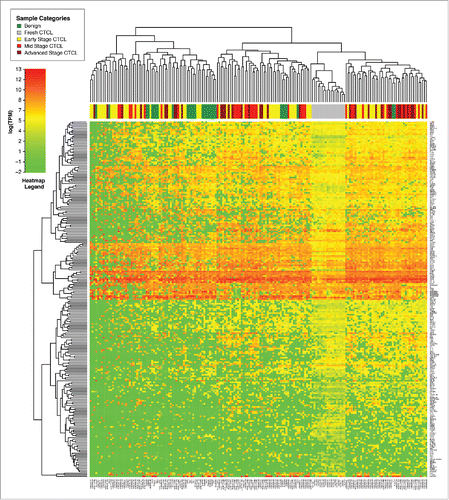
Table 2. Genes with statistically significant differences (based on Welch's t-test) in expression between benign skin lesions vs. all CTCL samples (left panel) and benign skin vs. stage IV CTCL samples (right panel). Average expression is presented as transcripts per million. p or q values for all analyses are reported. The q values that failed to reach statistical significance are presented in light grey font.
Figure 2. Unsupervised clustering analysis based on TruSeq targeted RNA gene expression of 284 select genes in benign (green) vs. stage IV (red) FFPE CTCL tissue samples.
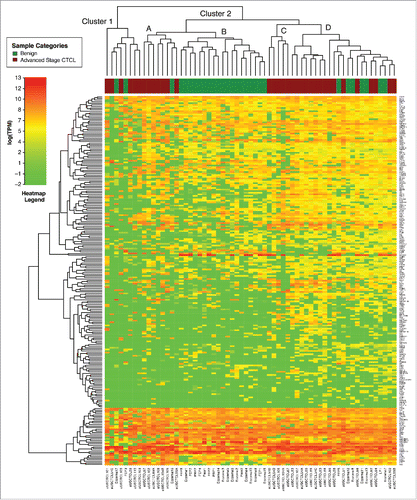
Figure 3. Unsupervised clustering analysis based on TruSeq targeted RNA gene expression of 284 select genes in early stage (≤IIA, yellow) vs. stage IIB and III (light red) vs. stage IV (dark red) FFPE CTCL tissue samples.
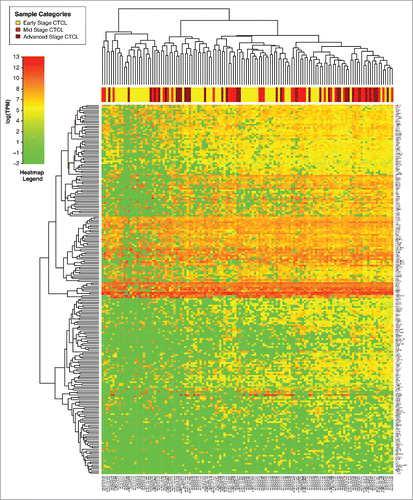
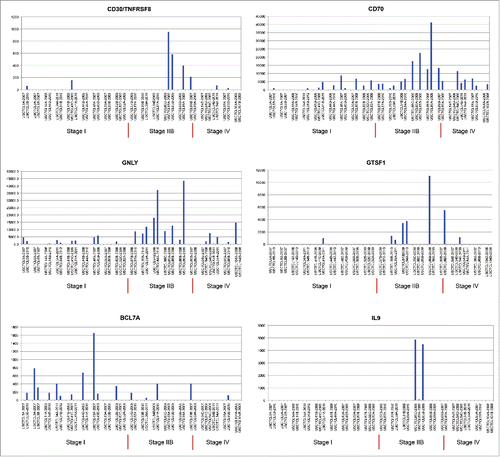
Table 3. (A) Genes with statistically significant differences in expression between early (≤IIa) vs. mid (staged IIB and III) and advanced (stage IV) CTCL samples (left panel). Results for the same gene presented for analysis of stage I vs. stage IV CTCL samples (right panel). While trends in gene expression were confirmed in both analyses only expression differences for GTSF1, LTBP4, IL18, CCL5, LCP2, TOX and FYB were statistically significant in both data sets. Expression differences for other genes in the latter analysis did not reach statistical significance due to lower number of samples in that set. The data that failed to reach statistical significance are presented in light grey font and p values for all analyses are reported. Average expression is presented as transcripts per million. (B) Genes with statistically significant differences in expression between stage I stable/indolent disease vs. stage I progressive disease.