Figures & data
Table 1. Characteristics of the subjects for whom cord blood was tested for maternal microchimerism.
Figure 1. Maternal microchimerism (MMc) data in cord blood mononuclear cells (CBMC) samples and by cellular subsets from healthy women. (A) Data from N = 51 CBMC samples are represented. The filled triangles represent 26 CBMC also selected for fluorescence-activated cell sorting into five cellular subsets in panel B. The open triangles are the remaining CBMC. (B) Data from N = 27 CBMC sorted into five cellular subsets. To help illustrate that measurements are not independent, each participating CB is represented by a slightly different shade of gray across the five subsets. Detection rate ratios (DRR), derived from the negative binomial regression model, are shown. DRR are interpreted as the fold-change in expected MMc quantities from one subset compared with a subset on its right. Numbers between brackets are 95% confidence intervals. Concentrations are measured in genome equivalent (gEq) of maternal DNA per 105 gEq of total cellular DNA and plotted in log scale. n/d = not detected.
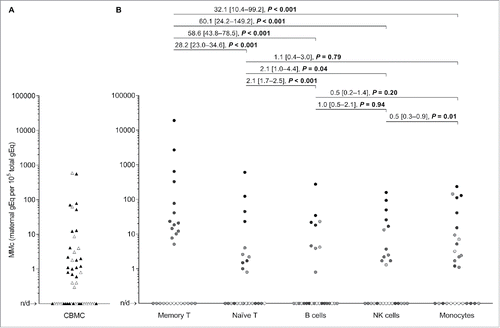
Figure 2. Maternal microchimerism (MMc) data by cellular subset, adjusted by the fraction of the corresponding subset in cord blood (CB) samples. To help illustrate the non-independence of measurements, each participating CB is represented by a slightly different shade of gray across the five subsets. Detection rate ratios (DRR), derived from the negative binomial regression model, are shown. DRR are interpreted as the fold-change in expected MMc quantities from one subset compared with a subset on its right. Numbers between brackets are 95% confidence intervals. Concentrations are measured in genome equivalent (gEq) of maternal DNA per 105 gEq of total cellular DNA and plotted in log scale. n/d = not detected.
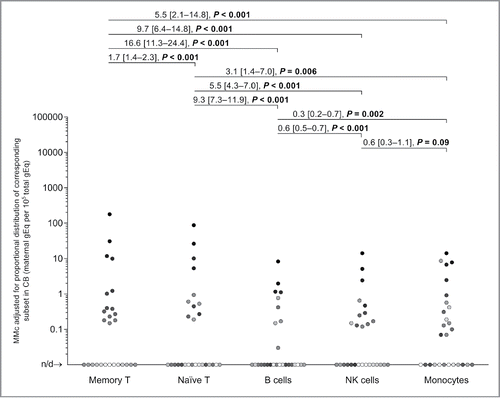
Figure 3. Pairwise correlation of maternal microchimerism (MMc) distribution in cellular subsets of cord blood (CB) and respective CB subjects. (A) The “matrix-like” figure represents the correlation between MMc levels in a particular subset vs. another subset for the same participating CB (each semi-transparent gray circle). Data is represented in genome equivalent (gEq) of maternal DNA per 105 gEq of total cellular DNA. n/d = not detected. (B) The heat maps show Pearson and Spearman correlation coefficients where blue, red, and white are negative (r tends to −1), positive (r tends to 1), and no (r = 0) correlation, respectively. p < 0.05 in all Pearson and Spearman's tests except in the two cases, where r = 0.31 and 0.33.
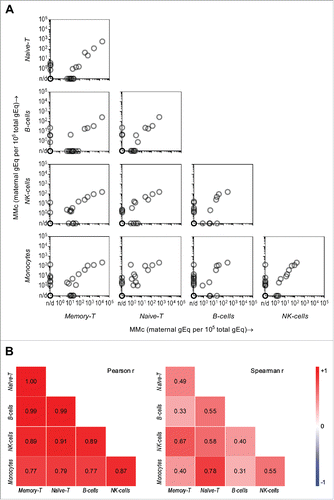
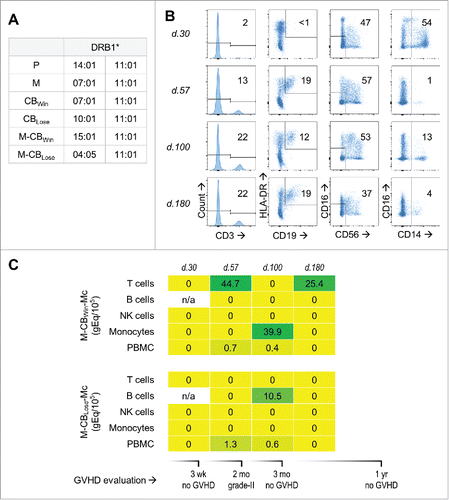
Figure 4. Maternal microchimerism (MMc) from the cord blood (CB) in cellular subsets of a leukemia patient who received double-CB transplantation. (A) HLA-DRB1 genotype of patient “P,” her mother “M,” the winning and losing received CB units: “CBWin” and “CBLose,” and the mothers of these CB: “M-CBWin” and “M-CBLose.” (B) From left to right, gating on T, B, NK cells, and monocytes sorted from patient's peripheral blood mononuclear cells (PBMC) at days 30, 57, 100, and 180 post-transplantation. Numbers in gates represent percentages. (C) Heat map revealing quantities of CB-origin MMc from winning (upper panel) and losing (lower panel) units at various time-points for various subsets, measured in genome equivalent (gEq) of maternal DNA per 105 gEq of total cellular DNA (gEq/105). Graft-vs.-host disease (GVHD) evaluation is also reported.