Figures & data
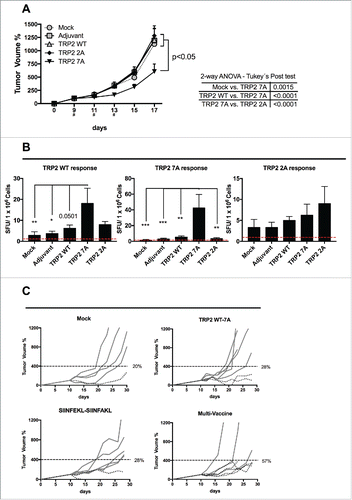
Figure 1. In silico screening of the mutational library of SIINFEKL. (A) Graphical representation of SIINFEKL peptide (violet) into the binding pocket of the H-2Kb MHC-I molecule (transparent gray). The backbone of the peptide is visualized in green and the lateral groups in blue. Lateral groups facing the MHC-I binding pocket or emerging from the pocket are indicated. In silico prediction of the binding affinity (B) or the immunogenicity score (C) of the analogs of SIIINFEKL. The analogs were grouped according to the position of the mutation (left panels). Alternatively, the analogs were grouped according to the aminoacid used to mutate each position (right panels). The red-dotted line, represents the IC50 and the immunogenicity score of the wild type sequence. (D) The affinity scores of the whole library were plotted and indicating analogs with a higher or lower affinity than the wild type SIINFEKL. The specific peptides SIINFAKL and SIIWFEKL are highlighted and their IC50 values are indicated. (E) The immunogenicity predictions for the H-2Kb allele are displayed for the wild type SIINFEKL and two specific analogs.
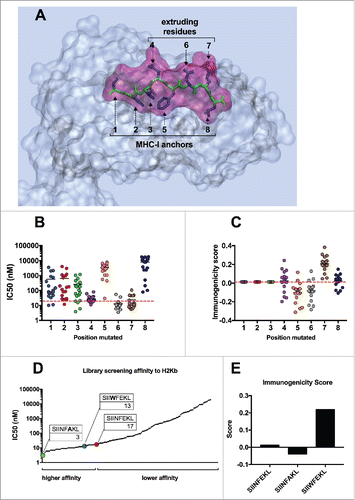
Figure 2. Molecular dynamics simulation to unravel conformational changes of epitope-MHC-I complexes. (A) Comparison of spatial conformation epitopes inside the MHC-I binding pocket. SIINFEKL (green transparent), SIINFAKL (red solid) and SIIWFEKL (cyan solid) are compared by superimposition. Molecular dynamics simulations were run for 300 ns, and the most representative states are shown. Left panel: SIINFEKL and SIINFAKL; central panel: SIINFEKL and SIIWFEKL; right panel SIINFAKL and SIIWFEKL. The conformational landscape of the epitope-MHC-I complexes (SIINFEKL, B; SIINFAKL, C; SIIWFEKL, D) are shown from two angles: C-terminal of epitope (left panels) and side views (right panels; epitopes are oriented from N-terminal on the left to C-terminal on the right). The extruding residues, responsible for contacting T-cell receptors are highlighted by yellow captions showing the position and abbreviation for the aminoacid. The structure and orientation of the mutated residue for SIINFAKL and SIIWFEKL peptides are highlighted by the yellow structure of the sidechains.
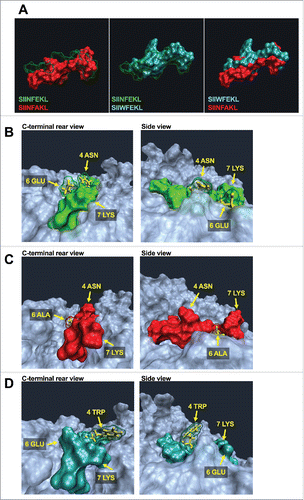
Figure 3. Experimental validation of in silico predictions of the MHC-I binding affinity. RMA-S cells were pre-incubated for 1 h at 4°C. Then, 4 × 106 cells were incubated for 2 h with one of the indicated peptides at two different concentrations (A) 1 µg /mL or (B) 0.1 µg/mL in a volume of 1 mL. The presence of H-2Kb molecules on the membrane was measured by flow cytometry and normalized against cells incubated with no peptide (negative control). (C) OT-I splenocytes were labeled with CFSE dye. Then, 3 × 104 cells were incubated with different stimuli for 72 h in a volume of 200 µL of complete media. Then the percentage of proliferating (i.e., CFSE diluted) CD3+CD8+ T-cells was determined by flow cytometry; data was normalized against positive control Concanavalin A (defined as 100% of proliferation). All the data are represented as mean ± SD; Significance was assessed by One-way ANOVA with Tukey's post hoc test; *p < 0.05, ** p <0.01.
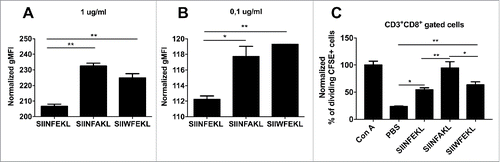
Figure 4. Antitumor response and in vivo efficacy of SIINFEKL mutated analogs. (A) B16OVA cells were injected into both flanks of female C57BL6/J mice (2.5 × 105 cells/flanks). Mice (eight per group) were treated with PBS (Mock), Ad5D24-CpG human adenovirus (Adjuvant), and either SIINFEKL, SIINFAKL or SIIWFEKL peptides complexed to the virus-adjuvant. Intra-tumor injections were made on day 9, 11, 13 as represented by the asterisks on the x axis. Tumor volumes were measured every 2–3 d by a digital caliper. Tumor volumes normalized against the values on the 9th day are presented as the mean ± SEM; significance was calculated by Two-way ANOVA with Tukey's post hoc test. Spleens, tumors and draining lymph nodes were collected to determine to percentage of (B) total CD8+ T-cells (CD19−CD3+CD8+) or (C) SIINFEKL-specific T-cells (double positive Pentamer+CD8+ gated on CD19−CD3+). (D) A correlation analysis was performed by plotting for each mouse the % of antigen-specific T-cells against the intensity of the signal (gMFI) of such population. Data from spleens (empty triangles), draining lymph nodes (empty squares) and tumors (black dots) are presented. Log transformed values were used to compute the Pearson's correlation coefficiencies; p-value for the correlation is 0.0002.
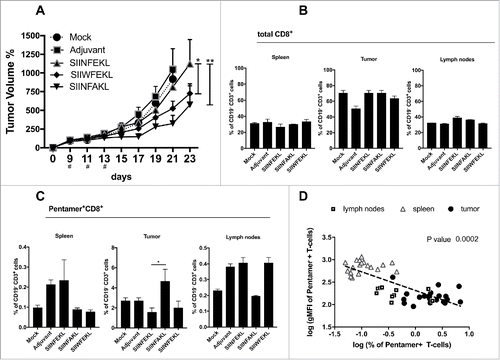
Figure 5. Cross-reactivity of the immunological responses elicited by the mutated analogs of SIINFEKL. Splenocytes from mice treated with SIINFEKL, SIINFAKL or SIIWFEKL (presented in , here indicated on the x axis), were assayed for cross-responses to the peptides. 3 × 105 cells were incubated with 100 ng of each indicated peptide for 72 h and the IFNγ response was determined by ELISPOT assay. (A) The number of Spot-Forming Units (SFU) is normalized to 1 × 106 cells. The red-dotted line represents the background of the assay (splenocytes incubated with PBS). (B) The cumulative size of the spots from mice of the same group (same column) in response to different stimuli (bars of different colors) is shown. Data is shown as mean ± SEM. C) The average spot size across all the groups in response to each peptide is represented. Data is shown as the mean ± SEM; One-way ANOVA with Tukey's post hoc test was used to calculate the statistical significance.
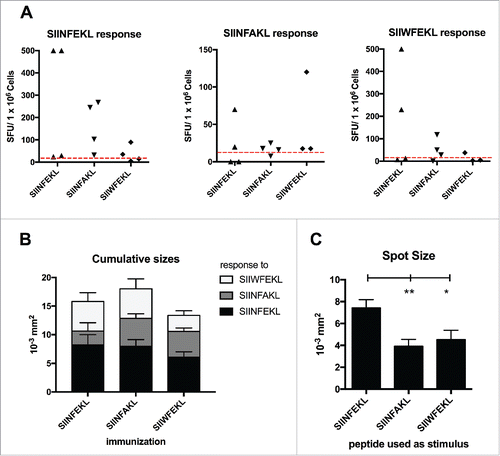
Figure 6. In silico study of the mutational library of the epitope TRP2180–188. The mutational library of the epitope SVYDFFVWL (TRP2 WT) was screened in silico for MHC-I binding affinity (A) or immunogenicity (B) for the allele H-2Kb. The analogs were grouped according to the position of the mutation (left panels) or according to the aminoacid used to mutate each position (right panels). C) The IC50 (left) and Immunogenicity score (right) of the analogs SVYDFFAWL (TRP2 7A) and SAYDFFVWL (TRP2 2A) predicted in silico are represented. (D) RMA-S cells were pre-incubated for 1 h at 4°C. Then, 4 × 106 cells were incubated for 2 h with one of the indicated peptides at two different concentrations 0.1 µg/mL (left) or 1 µg/mL (right) in a volume of 1 mL. The presence of H-2Kb molecules on the membrane was measured by flow cytometry and normalized against cells incubated with no peptide (negative control). Data is represented as the mean ± SEM; Unpaired student's t-test, *p < 0.05, **p < 0.01. E) Comparison of spatial conformation epitopes inside the MHC-I binding pocket: TRP2 WT (orange), TRP2 7A (green) and TRP2 2A (purple). Molecular dynamics simulations were run for 300 ns, and the most representative states are shown. ¾ view (left panel) and side view (right panel). The mutated residues are colored in red. (F) The conformational landscapes of the epitope-MHC-I complexes are shown from peptide C-terminal perspective. The extruding residues, responsible for contacting T-cell receptors are highlighted by yellow captions showing the position and abbreviation for the aminoacid. The α chain of the MHC is represented in transparent white.
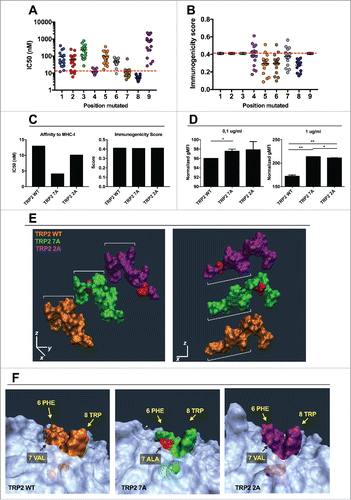
Figure 7. Breaking the tolerance to TRP2 tumor antigen and evaluation of multivaccine therapy. (A) B16-F10 melanoma cells were injected into both flanks of female C57BL6/J mice (1.5 × 105 cells/flank). Mice (eight per group) were treated with PBS (Mock), Ad5D24-CpG human adenovirus (Adjuvant), and either TRP2 WT, TRP2 7A or TRP2 2A peptides complexed to the virus-adjuvant. Intra-tumor injections were made on day 9, 11 and 13 (asterisks on the x axis). Tumor volumes normalized against the values on the 9th day and are presented as the mean ± SEM (data from two independent experiments); significance was calculated by Two-way ANOVA with Tukey's post hoc test. (B) Splenocytes were collected from mice and they were assayed for cross-responses to the peptides. 4 × 105 cells were incubated with 100 ng of indicated stimuli for 72 h and the IFNγ response was determined by ELISPOT assay. The number of Spot-Forming Units (SFU) is normalized to 1 × 106 cells. The red-dotted line represents the background of the assay (splenocytes incubated with PBS). Data is presented as mean ± SEM; Significance was calculated by using One-way ANOVA with Tukey's post hoc test; *p < 0.05,**p < 0.01, ***p < 0.001. (C) B16OVA bearing mice were treated on day 10, 12 and 14 with PBS (mock), a combination of TRP2 WT and 7A peptides, a combination of SIINFEKL and SIINFAKL peptides and a multitherapy featuring all four different epitopes together. Tumor volumes normalized against the values on the 10th day. A threshold of 400% of initial volume on day 10 was chosen to distinguish between fast (solid line) and slow (dotted line) progressing tumors. Percentages of slow progressing tumors are indicated near the graph of each group.