Figures & data
Figure 1. Platelet-induced shedding of NKG2DL impairs NK antitumor reactivity. The indicated tumor cells were cultured for 48 h alone or in the presence of platelets (ratio tumor cells/platelets of 1:150) to enable coating. After washing to remove surplus platelets and soluble factors, (A) surface expression of MICA, MICB, ULBP1–3 and EGFR/CD133 as control were analyzed by flow cytometry. Open peaks, tumor cells alone; shaded peaks, platelet-coated tumor cells; dotted lines, isotype controls. (B) Levels of sNKG2DL in supernatants were analyzed by ELISA. Statistically significantly different results are indicated by #. (C) Tumor cells were incubated with pNKC in the presence or absence of anti-NKG2D F(ab´)2 fragments or isotype control (both 5 µg/ml). Target cell lysis was determined by 4 h chromium release assays. Results obtained at the indicated E:T ratios (left) and the calculated reduction of lysis (δ) by NKG2D blockade (at the lowest E:T ratio, right) are displayed. Representative data of one experiment from a total of at least 4 with similar results are shown.
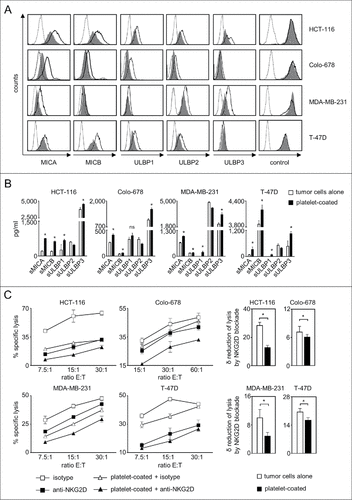
Figure 2. ADAM10 and ADAM17 expression in thrombopoietic cells. (A) CD34+ haematopoietic progenitor cells were cultured for 12 d in the presence of thrombopoietin to induce megakaryocyte differentiation. On days 0, 6, 9, and 12 mRNA expression of ADAM10 and ADAM17 was analyzed by qPCR. Representative data of one from a total of 3 biologic experiments performed in technical replicates are shown. (B) Platelets from healthy donors were either left untreated (resting) or activated with ADP, collagen or thrombin until aggregation was visible (for a maximum of 30 s to prevent clumping) as described previously.Citation23 Then FACS analysis of ADAM10 or ADAM17 expression was performed. (C) Platelets were either left untreated (resting) or activated by exposure to the platelet superagonist TRAP-6 (5 µM) for 5 minutes. Then ADAM10 and ADAM17 levels in platelet pellets and releasate was analyzed by Western blot. In (B-C), representative data of one experiment from a total of at least 4 with similar results are shown. (D) Resting platelets from healthy donors and patients with NSCLC were analyzed by FACS as described in (B). Statistically significant differences (p<0.05, Kolmogorov-Smirnov test) are indicated by #.
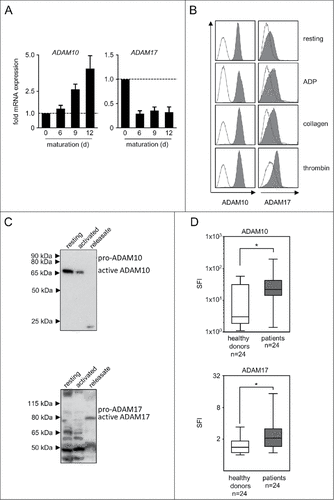
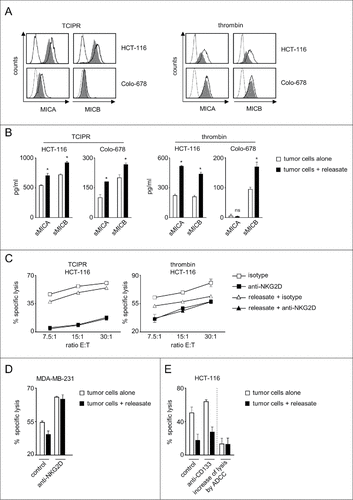
Figure 3. Platelet-releasate mediates shedding of NKG2DL and impairs NK cytotoxicity. The indicated tumor cells were cultured for 48 h alone or in the presence of thrombin-induced releasate or TCIPR generated by activation of platelets with the same tumor cells that were subsequently treated with the respective releasate. Then supernatants were collected and tumor cells were washed to remove surplus platelets and soluble factors. Subsequently (A) surface expression of MICA and MICB on tumor cells was analyzed by flow cytometry. Open peaks, tumor cells alone; shaded peaks, tumor cells cultured with releasate; dotted lines, isotype controls; (B) levels of sMICA and sMICB in supernatants were analyzed by ELISA. Statistically significantly different results are indicated by #. (C) The indicated tumor cells were incubated with pNKC in the presence or absence of anti-NKG2D F(ab´)2 fragments or isotype control (both 5 µg/ml). Target cell lysis was determined by 4 h chromium release assays. (D) NK cell lysis of tumor cells (E:T ratio 3.75:1) was analyzed in the presence of absence of an agonistic NKG2D antibody or isotype control (both at 10 µg/ml) as described in C. (E) NK lysis of tumor cells in the presence or absence of an Fc-optimized CD133 antibody or isotype control (10 µg/ml each) was determined as described in C. Results obtained at an E:T ratio 7.5:1 and the calculated increase of lysis as induced by ADCC are shown. Representative data of one experiment from a total of at least 4 with similar results are shown.