Figures & data
Figure 1. Infiltration of tumors by T cells and T cell subsets. (A) The T cell infiltration score was calculated by evaluating the mRNA expression of genes included in the T cell signature proposed by Bindea et al.Citation31 However, the genes TRA and TRAC are not included in the Affymetrix platform and therefore were not considered. The ratio of the expression in the patient's tumor sample and the mean expression in the HTs was calculated for each gene. Then, the mean ratio of the genes in each patient was expressed as log(2). The mean normalized expression of genes included in the T cell signature in tumors T1, T2, T3 and T4 was found to be significantly higher as compared to T6, T7, T8 and T9 (####, p<0.0001). (B) The CTL activity score was calculated by evaluating the mRNA expression of genes included in the CTL signatureCitation31 normalized to the expression of CD8α. The ratio of the expression in the patient's tumor sample and the mean expression in the PBMCs of HDs was calculated for each gene. Then, the mean ratio of the genes in each patient was expressed as log(2). The mean normalized expression of genes included in the CTL signature in each tumor was found to be significantly lower (##, p<0.01; ###, p<0.001; ####, p<0.0001) or not significantly different (n.s.) as compared to the mean normalized expression in PBMCs of HDs. (C) The suppressive score was calculated by evaluating FOXP3, IL-10 and TGFβ mRNA expression normalized to the expression of CD4. The ratio of the expression in the patient's tumor sample and the mean expression in the PBMCs of HDs was calculated for each gene. Then, the mean ratio of the genes in each patient was expressed as log(2). The mean normalized expression of FOXP3, IL-10 and TGFβ in each tumor was found to be significantly higher as compared to the mean normalized expression in PBMCs of HDs (####, p<0.0001). (D) Correlation of TGFβ mRNA expression and CD4 mRNA expression in patients with non-squamous NSCLC. The Spearman coefficient (ρ) and p value are reported. (E) Correlation of IL-10 mRNA expression and CD4 mRNA expression in patients with renal cell carcinoma. Spearman coefficient (ρ) and p value are reported. All box-and-whisker plots (A-C) represent values within the interquartile range (boxes), and whiskers represent the highest and the lowest values.
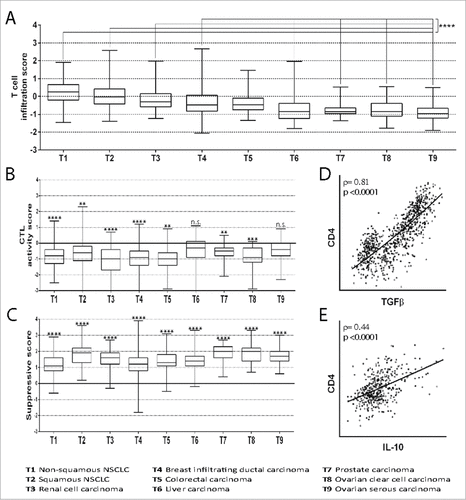
Table 1. Decrease in the CD8+/CD4+ ratio in cancer specimens as compared to PBMCs from HDs.
Table 2. Correlation between mRNA expression of the regulatory cytokines and the mRNA expression of CD4 in cancer specimens.
Table 3. Correlation between the mean mRNA expression of genes representing T cells (see Figure S1) and the mean mRNA expression of genes representing CTL, normalized by CD8α expression, in cancer specimens.
Figure 2. Expression of Treg markers in tumors vs. HTs. The ratio of expression of the marker in the tumor to its expression in HTs is shown on a log(2) scale. Markers with higher than two-fold expression in the tumor compared to the corresponding HTs (#, p<0.05) or higher than 1.5-fold expression in the tumor compared to both the corresponding HTs (#, p<0.05, as evaluated by Student's t-test) and the mean expression in HTs (#, p<0.05) were selected for further analysis (markers within grey boxes). As a rescue strategy (prostate carcinoma), markers showing higher than 1.5-fold expression in the tumor as compared to the corresponding HTs (#, p<0.05) were selected for further analysis (marker within grey dotted boxes). Markers that met the expression conditions but were not selected because the difference between tumors and HTs was not significant (n.s.) are shown within white boxes.
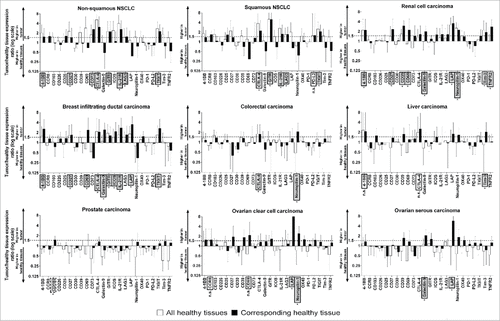
Table 4. Overexpression of Treg markers in specific tumors.
Figure 3. mRNA expression of the best Treg marker in tumor samples and the corresponding HTs. Mean±SD values of expression of the Treg marker in HTs and expression of the marker in each tumor sample are shown. The percentage of patients with expression greater than the mean+2SD value (dotted line) of the corresponding HTs is reported. The best Treg marker for each tumor is shown (the expression of the other selected Treg markers are shown in supplemental Figures S2−S7). Student's t-test was used for statistical analysis in the comparison between the expression level of the Treg marker mRNA in tumors vs. the corresponding HT samples.
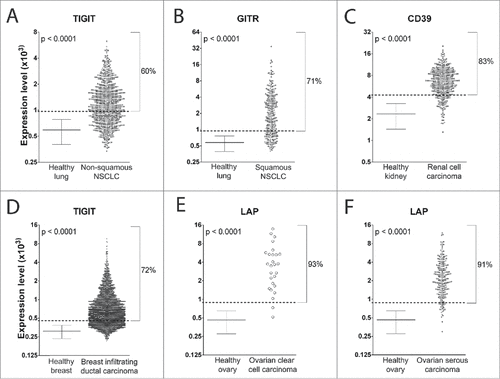
Figure 4. Correlation between the expression of GITR and suppressive cytokines. Correlation of GITR mRNA expression with TGFβ (A) and IL-10 (B) mRNA expression in patients with non-squamous NSCLC. Spearman coefficients (ρ) and p values are reported.
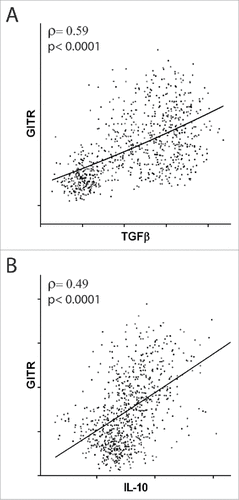
Table 5. Correlation of the expression of Treg markers with that of the suppressive cytokines in tumor samples.
Figure 5. Expression of activating FcγR mRNA in tumors vs. lymph nodes from HDs. The ratio of the mRNA expression of the specified activating FcγR in the tumor specimen and the mean (±SD) mRNA expression in the lymph nodes of HDs is shown on a log(2) scale. In the histogram on the right, the mean of the ratios [expressed as log(2)] of the four activating FcγRs is shown. The mean expression of each FcγR in each tumor as compared to the mean expression in HD lymph nodes was found to be significantly different (#, p<0.05: ##, p<0.01: ###, p<0.001; ####, p<0.0001).
![Figure 5. Expression of activating FcγR mRNA in tumors vs. lymph nodes from HDs. The ratio of the mRNA expression of the specified activating FcγR in the tumor specimen and the mean (±SD) mRNA expression in the lymph nodes of HDs is shown on a log(2) scale. In the histogram on the right, the mean of the ratios [expressed as log(2)] of the four activating FcγRs is shown. The mean expression of each FcγR in each tumor as compared to the mean expression in HD lymph nodes was found to be significantly different (#, p<0.05: ##, p<0.01: ###, p<0.001; ####, p<0.0001).](/cms/asset/031af5d5-b9e6-4f91-b26f-e50d4646ca08/koni_a_1387705_f0005_b.gif)
Figure 6. Algorithm used to calculate the ADCC index of Treg markers in a cancer. To evaluate the probability of a Treg marker being successfully used as a target to induce ADCC, we established the ADCC index. The ADCC index was calculated for each marker in each cancer as described in the material and methods. Half of the score is derived from evaluation of overexpression of the marker in the responder patients (i.e., the patients in whom the Treg marker expression level was higher than the mean+2SD value of the Treg marker expression in HTs), one-fourth from the expression of activating FcγR in the cancer sample of the responder patients, and one-fourth from the correlation of the marker expression with the suppressive microenvironment (i.e., the probability that the marker was mainly expressed in Tregs). The final score was obtained by multiplying each of these four scores. The ADCC scores of the markers in specific cancers are shown in .
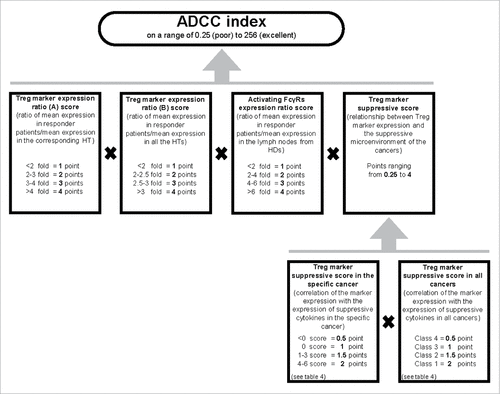
Table 6. ADCC index of the markers overexpressed in more than 40% of the patients.
