Figures & data
Table 1. Characteristics of cell lines established from metastatic melanoma.
Figure 1. Cell surface expression of MHC, NKG2D-ligands and DNAM1-ligands on melanoma cells. The metastatic melanoma cells Ma-Mel-55 (55), Ma-Mel-86c (86c), Ma-Mel-86f (86f) and Ma-Mel-103b (103b) were stained with the indicated antibodies and analysed by flow cytometry. Isotype control appears in grey, and signal for each antibody as a black line. The panel shows one representative experiment out of 3–5.
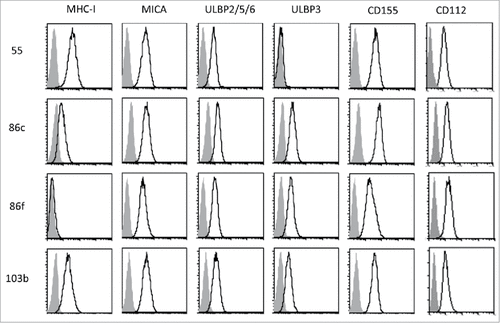
Figure 2. Effect of vemurafenib on NK ligand expression in melanoma cells. Melanoma cells Ma-Mel-55 (55), Ma-Mel-86c (86c), Ma-Mel-86f (86f) and Ma-Mel-103b (103b) were treated with 1 μM vemurafenib (PLX) for 48 h (A,B, D) or 24 h (C) (control cells were treated with the carrier DMSO) A. Flow cytometry. Melanoma cells were stained for detection of the indicated markers by flow cytometry. The plots represent the change in the mean fluorescence intensity (MFI) of each marker, as the percentage of the molecule present in control (DMSO) cells. Data are the mean and SEM. Isotype MFI was subtracted (n≥3, a representative experiment is shown in Suppl. ). B. Western blot showing the total amount of MICA in whole cell lysates, using antigen affinity-purified biotinylated goat polyclonal anti-MICA antibody BAF1300. Actin was used as loading control (n = 4). C. Soluble MICA and ULBP2/5/6 released to supernatants of vemurafenib -treated metastatic melanoma cells was analysed by ELISA at 24 h post-treatment. Plots represent the mean and SEM of protein concentration (ng/ml) (n = 3). D. mRNA detected using qPCR. Cells were recovered to extract RNA. cDNA was prepared and used as template in qRT-PCR experiments. Data are the mean and SEM, relative to control (DMSO) cells. RPLP0 mRNA levels were determined for normalization (#p < 0.05 ##p < 0.01, ### p < 0.001, #### p < 0.0001).
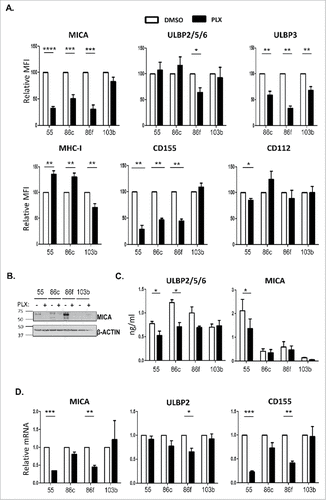
Figure 3. Kinetics of the effect of vemurafenib on surface NK ligand expression and proliferation in melanoma cells. A. Effect of vemurafenib on NKG2D-ligand surface expression. Metastatic melanoma cells Ma-Mel-55 (55), Ma-Mel-86c (86c), Ma-Mel-86f (86f) and Ma-Mel-103b (103b) were treated with 1 μM vemurafenib for either 1 h, 24 h or 48 h (control cells with the carrier DMSO) and stained for detection of the indicated markers by flow cytometry. The plots represent the change in the mean fluorescence intensity (MFI) of each marker, as the percentage of the molecule present in control (DMSO) cells. Data show the mean and SEM. Isotype MFI was substracted (#p < 0.05 ##p < 0.01, ### p < 0.001) (n = 3). B. Proliferation assay. Metastatic melanoma cells were allowed to adhere to E-plates and then treated with 1 μM vemurafenib (control cells with the carrier DMSO). Real time electrical impedance was monitored to quantify cell proliferation for 48 h using x-CELLigence. Cell index normalized to the treatment time is represented. Arrows indicate the time of vemurafenib addition (PLX), 24 and 48 h post-treatment. The plots show quadruplicates of one representative of 3 independent experiments.
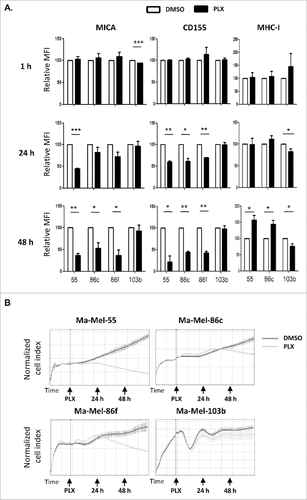
Figure 4. Effect of vemurafenib on NK cell recognition of melanoma cells. Metastatic melanoma cells Ma-Mel-55 (55), Ma-Mel-86c (86c), Ma-Mel-86f (86f) and Ma-Mel-103b (103b) were treated 48 h with either 1 μM vemurafenib or DMSO and used as targets in experiments of degranulation using NK cells. The plots represent the percentage of NK cells positive for surface CD107a (LAMP1) as a measure of degranulation. A. Healthy donor NK cells. Each symbol corresponds to an experiment with a different donor (n = 4). B. Autologous NK cells. Ma-Mel-86f were used as target cells. Autologous NK cell were also treated 48 h with DMSO or vemurafenib. Data represent the relative decrease as percentage of untreated cells. Three independent experiments are shown (n.s., non-significant; #p < 0.05 ## p < 0.01).
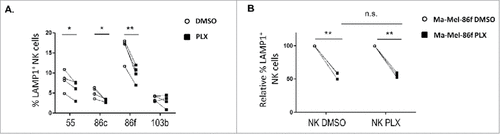
Figure 5. Effect of the combination of HDACi and BRAFi on the immunophenotype and proliferation of melanoma cells. A. Titration of sodium butyrate and surface expression of NK ligands. Metastatic melanoma cells Ma-Mel-55 (55), Ma- Mel-86c (86c) and Ma-Mel-103b (103b) were treated with the indicated amounts of sodium butyrate or water for 18 h and stained with antibodies for detection of the indicated markers by flow cytometry. The plots represent the change in the mean fluorescence intensity (MFI) of each marker, as the percentage of the molecule present in control (water) cells. Data show the mean and SEM corresponding to 3 experiments. B. MTT assay of melanoma cells treated with the drug combination. Metastatic melanoma cells Ma-Mel-55 (55), Ma- Mel-86c (86c), Ma-Mel-86f (86f) and Ma-Mel-103b (103b) were treated with either 1 μM vemurafenib, 5 mM sodium butyrate or the combination of both for 24 h (control cells with the carrier, DMSO and water, CTRL). Cells were assayed for metabolic activity by incubation with MTT and OD570 was measured. Data represented are the mean as percentage of untreated cells and SEM of four replicates, from one experiment out of two. C. NK ligands on melanoma cells treated with the drug combination. Cells were stained for detection of the indicated markers by flow cytometry. The plots represent the mean fluorescence intensity (MFI) of each marker, isotype subtracted and the relative change as the percentage of the molecule present in control (DMSO and water, CTRL) cells. Data show the mean and SEM corresponding to 3 experiments. D. mRNA detected using qPCR. Cells were recovered to extract RNA. cDNA was prepared and used as template in qRT-PCR experiments. RPLP0 mRNA levels were determined for normalization (#p < 0.05 ##p < 0.01, ### p < 0.001). For simplicity, in B, C and D, only the p-values of each inhibitor related to the combination are shown.
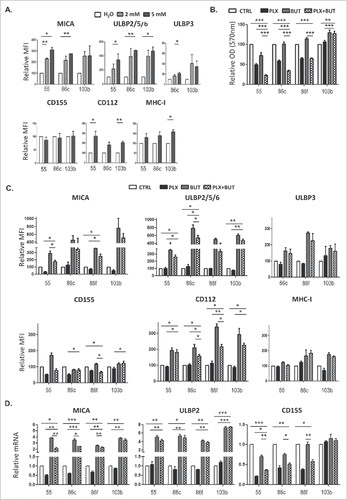
Figure 6. A. NK cell degranulation in the presence of HDACi- and BRAFi-treated melanoma cells. Melanoma cells Ma-Mel-55, Ma-Mel-86c, Ma-Mel-86f and Ma-Mel-103b cells treated with either 1 μM vemurafenib, 5 mM sodium butyrate or the combination of both for 24 h (control cells with the carrier, DMSO and water, CTRL) were used as targets in experiments of degranulation using NK cells from healthy donors PBMCs. The plots represent the percentage of NK cells positive for surface CD107a (LAMP1) as a measure of degranulation in 4 experiments (#p < 0.05 ##p < 0.01). For simplicity, only the p-values of each inhibitor related to the combination are shown. B. Effect of the blockade of activating NK receptors in degranulation experiments in the presence of HDACi- and BRAFi-treated melanoma cells. Melanoma cells Ma-Mel-86c treated with carrier (either DMSO or water, control) and the combination of 1 μM vemurafenib and 5 mM sodium butyrate for 24 h were used as targets in experiments of degranulation using NK cells from healthy donors PBMCs. NK receptors (NKG2D, DNAM-1, or both) were blocked, as indicated, incubating PBMCs with the corresponding antibodies, or isotype (ISO) as negative control. The plots represent the percentage of NK cells positive for surface CD107a (LAMP1) as a measure of degranulation in 3 experiments (#p < 0.05 ##p < 0.01###p < 0.001).
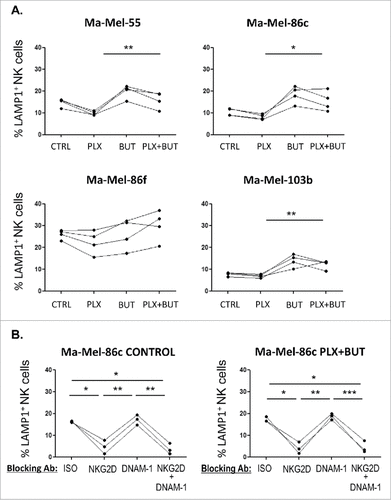
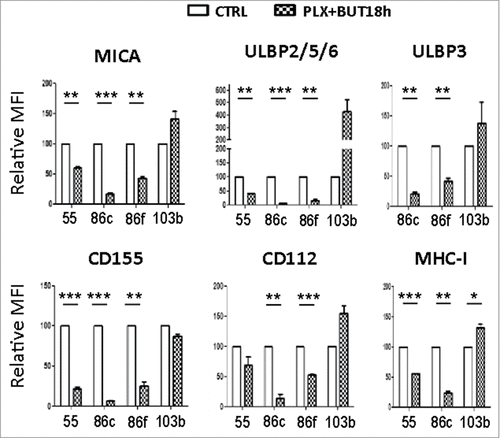
Figure 7. NK ligand expression on melanoma cells sequentially treated with vemurafenib (48 h) and sodium butyrate (last 18 h). Melanoma cells Ma-Mel-55 (55), Ma-Mel-86c (86c), Ma-Mel-86f (86f) and Ma-Mel-103b (103b) were treated with 1 μM vemurafenib (PLX) for 48 h, for the last 18 h of culture, a final concentration of 5 mM sodium butyrate was added (control cells with the carrier DMSO or water). Cells were stained for detection of the indicated markers by flow cytometry. The plots represent the mean fluorescence intensity (MFI) of each marker, isotype subtracted and the relative change as the percentage of the molecule present in control (DMSO and water, CTRL) cells. Data show the mean and SEM corresponding to 3 experiments. (#p < 0.05 ##p < 0.01, ### p < 0.001).