Figures & data
Figure 1. Phenotypic analysis of PDA immune infiltrating cells. (A) Characterization of T cells and F4/80 positive and negative populations in normal pancreas (NP) versus PDA. PDL-1 expression on F4/80hi and F4/80low cells is shown as delta Geo Mean of the fluorescence intensity after subtraction of isotype-related values. (B) Analysis of cytokine production and surface marker expression in NP or PDA-infiltrating T cells. Each graph shows the percentage of positive cells for the specific marker as indicated on the Y-axis. Data are represented as mean ± SEM of pooled cells from three independent experiments with two/three mice per experiment. Statistical analysis by unpaired Student's t test.
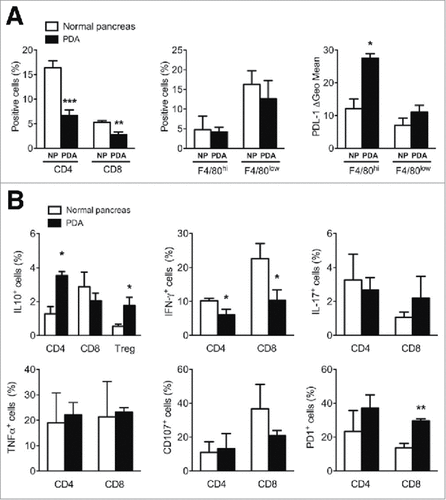
Figure 2. Epigenetic changes of Il10 and T-bet promoters in PDA-infiltrating T cells. (A) ChIP analysis targeting H3K4me3 and H3K27me3 at Il10 promoter in CD4, CD8 and Treg cells sorted from NP and PDA. (B) ChIP analysis targeting H3K4me3 and H3K27me3 at T-bet promoter in CD4 and CD8 T cells sorted from NP and PDA. Columns represent the percentage of input chromatin. Data are represented as mean ± SEM of pooled cells from two independent experiments with three mice per experiment, in which tumor cell suspensions were pooled together for the sorting. Statistical analysis by unpaired Student's t test. #, ##, ### P values statistically different between NP and PDA; §§§ P values statistically different between permissive and repressive marks in the NP group.
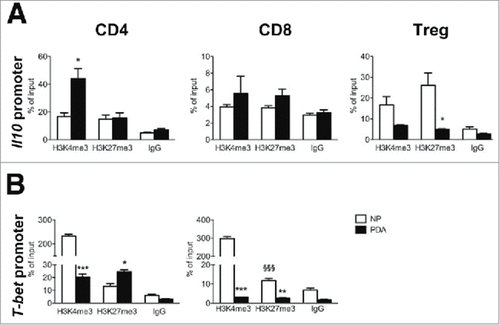
Figure 3. Phenotypic analysis of the PDA microenvironment after Trabectedin treatment. (A) Left panel: PDA wet weight from untreated (NT) and Trabectedin-treated (Tra) mice. Data are represented as box and whiskers of Tukey's method (six mice per group; statistical analysis by unpaired Student's t test). Right panel: Flow cytometry analysis of Monocytes (M∅) and Polymorphonuclear cells (PMN) in blood collected 24 h after each Trabectedin treatment from NT and Trabectedin-treated mice. (B) Absolute number of infiltrating immune cells positive for specific markers, expressed per 104 leukocytes from NT and Trabectedin-treated mice. (C) Analysis of PDA-infiltrating T cell cytokine production and surface markers from NT or Trabectedin-treated mice. Each plot shows the percentage of positive cells for the specific marker, as indicated on the Y-axis. Data are represented as means ± SEM of pooled cells from two independent experiments with three mice per condition. Statistical analysis by unpaired Student's t test.
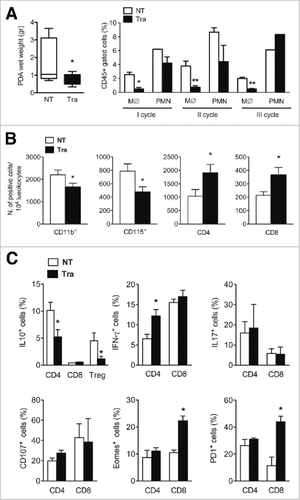
Figure 4. Epigenetic changes of Il10 and T-bet promoters in PDA-infiltrating T cells after Trabectedin treatment. ChIP analysis targeting H3K4me3 and H3K27me3 of (A) Il10 and (B) T-bet promoters in PDA-infiltrating CD4 and CD8 T cells sorted from NT and Trabectedin-treated mice. Columns represent percentage of input chromatin. Data are represented as means ± SEM of pooled cells from two independent experiments, in which tumor cell suspensions from three mice were pooled together for the sorting. Statistical analysis by unpaired Student's t test. #, ## P values statistically different between NT and Tra; §, §§ P values statistically different between permissive and repressive marks in the NT or Tra group.
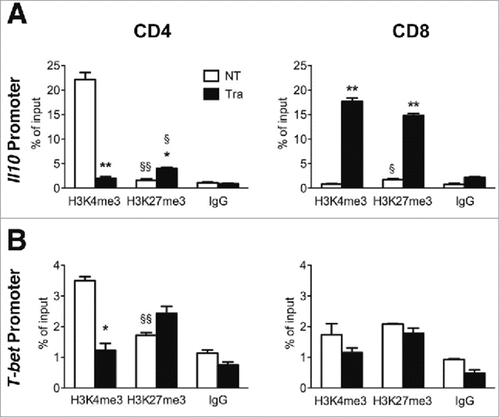
Figure 5. Effects of Trabectedin on CD4 T cells. ChIP analysis of H3K4me3 and H3K27me3 of Il10 (A) and T-bet (B) and Pdcd1 promoter (C) and associated enhancers at −3.7kb and +17.1kb from the transcription start site (D) in sorted CD4 T cells from naïve mice alone or in the presence of Trabectedin and stimulated with supernatants from M∅ in the presence or absence of Trabectedin. Columns represent percentage of input chromatin. Data are represented as means ± SEM of pooled cells from two independent experiments. Statistical analysis by unpaired Student's t test. #, ##, ### P values statistically different between NT and Tra; §, §§ P values statistically different between permissive and repressive marks in the NT or Tra group.
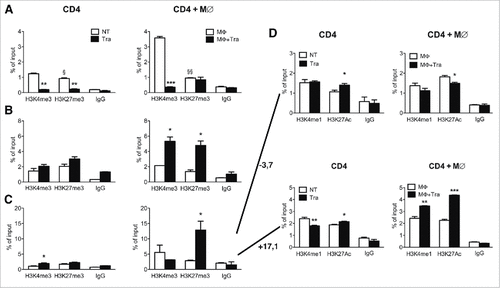
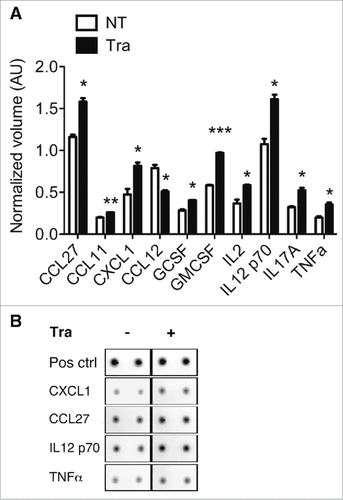
Figure 6. Evaluation of chemokine/cytokine secretion by macrophages after Trabectedin treatment. (A) Quantification of cytokines and chemokines in the supernatants of macrophages treated with Trabectedin for 24h, or untreated. Data are represented as means of normalized volumes (arbitrary units – AU) ± SEM from two independent experiments. Statistical analysis by unpaired Student's t test. (B) Representative spots from the recorded images of one array.