Figures & data
Figure 1. Characterization of immune infiltration in E0771 TNBC tumor model. (A) E0771 cells were orthotopically implanted into the mammary fat pad of C57 BL/6 mice and tumor volume was measured biweekly. (B) Tumors were enzymatically digested and analyzed for immune cell infiltration when tumors reached terminal endpoint volume. Gating as follows: CD45+CD11-CD4+ or CD8+ T cells, CD45+CD11b+Ly6ChiLy6G- monocytes, CD45+CD11b+Ly6Ghi neutrophils, CD45+CD11b+Ly6 C-Ly6G-F480+ macrophages (C) CD45+CD11b-CD8β-CD4+FoxP3+ Tregs from spleens and tumors were quantified by flow cytometry when tumors reached terminal endpoint volume. (D) Representative histograms from tumors and spleens pregated on live, CD45+, CD11b-, CD8β-, CD4+, FoxP3+ or FoxP3- (top) and quantification (bottom). (E) CD45+CD11b-CD8β+ or CD45+CD11b-CD4+ cells from the spleen or tumors were analyzed for expression of PD1 when tumors reached terminal volume. (F) E0771 cells grown in culture, E0771 cells grown in culture with 10ng/ml recombinant IFNγ for 48 hours, or taken ex vivo from tumors grown orthotopically in the mammary fat pad of C57 BL/6 mice when tumors reached terminal endpoint volume were stained for PDL1 expression. Error bars indicate SEM. Representative of 3 experiments. n = 5 mice per group; *P < 0.05; **P < 0.01; ***P < 0.001 by paired t-test.

Figure 2. Dual blockade of PD1 and CTLA4 has profound anti-tumor effect on E0771 TNBC tumors and TCR repertoire. (A) E0771 cells were orthotopically implanted into the mammary fat pad of C57 BL/6 mice and measured biweekly. Mice were randomized into groups and treated with anti-PD1 and/or anti-CTLA4 (IgG2 a) antibodies biweekly beginning when tumors measured >100 mmCitation3 or were treated with isotope controls antibodies. (B-D) High-throughput quantitative sequencing of the rearranged TCR β genes of tumor or PBMC samples. Analyses were performed using immunoSEQ analyzer software (Adaptive Biotechnologies) and represent a single experiment. (B) Number of total productive TCR templates present in tumors. (C) Clonality score of TCRs present in tumors. (D) Number of unique TCR rearrangements present in tumors. (E) Frequency of the top ten TCR clones found in individual tumor samples. Each color (red through green) at the top of the bars represents the top 10 individual clones. The blue bar represents all remaining clones present in the sample. (F-H) Similarity heat map between individual tumor and PBMC samples. Dark red score of 1 is exactly the same and white score of 0 is completely dissimilar. (F) Similarity between tumor samples. (G) Similarity between PBMC samples. (H) Similarity between tumor and PBMC samples. Error bars indicate SEM. n = 5 per group for A; n = 3-5 per group for B-E; n = 2-5 per group for F; *P < 0.05; **P < 0.01, ***P < 0.001 by one-way ANOVA with Bonferroni's multiple comparisons test to isotype control group.
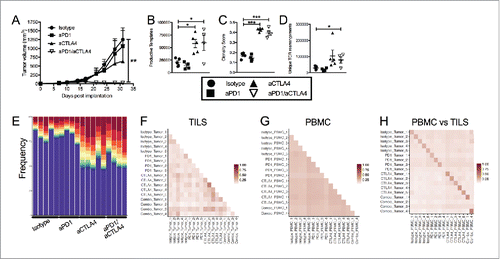
Figure 3. Dual blockade of PD1 and CTLA4 has profound anti-tumor effect on neoantigen expressing E0771-OVA tumors and has same effect on TCR repertoire. (A) E0771-OVA cells were grown subcutaneously in C57 BL/6 mice and tumor volume was measured biweekly. Mice were randomized into groups and treated with anti-PD1 and/or anti-CTLA4-IgG2 a antibodies biweekly beginning when tumors measured >100 mmCitation3 or left untreated. (B) Splenocytes from mice in (A) taken at the terminal endpoint were stimulated as indicated and IFN-γ producing cells were analyzed by ELISPOT. (C) Serum from mice in (A) taken at the terminal endpoint was analyzed by ELISA for anti-OVA antibodies. (D) CD4+ and CD8+ T cells from tumors of mice in (A) taken at the terminal endpoint were quantified by flow cytometry. (E) CD4+ FoxP3+ Tregs from tumors of mice in (A) taken at the terminal endpoint were quantified by flow cytometry. (F) Effector T cell: Treg ratio from (D) and (E) was calculated. A-E representative of 3 experiments. (G-M) High-throughput quantitative sequencing of the rearranged TCR β genes. Analyses were performed using immunoSEQ analyzer software (Adaptive Biotechnologies) and represent a single experiment. (G) Number of total productive TCR templates present in tumors. (H) Clonality score of TCRs present in tumors. (I) Number of unique TCR rearrangements present in tumors. (J) Frequency of the top ten TCR clones found in individual tumor samples. Each color (red through green) at the top of the bars represents the top 10 individual clones. The blue bar represents all remaining clones present in the sample. (K) Frequency of the top clone (left) and the OTI TCR clone (right) in each E0771-OVA tumor (L) Frequency of the top clone in each E0771 tumor (M) Similarity heat map between individual tumor samples. Dark red score of 1 is exactly the same and white score of 0 is completely dissimilar. Error bars indicate SEM. n = 5 per group for A-F; n = 3-5 per group for G-M; *P < 0.05; **P < 0.01; ***P < 0.001 by one-way ANOVA with Bonferroni's multiple comparisons test to E0771-OVA control group; P value in (C) are for both aCTLA4 and aCTLA4/aPD1 groups.
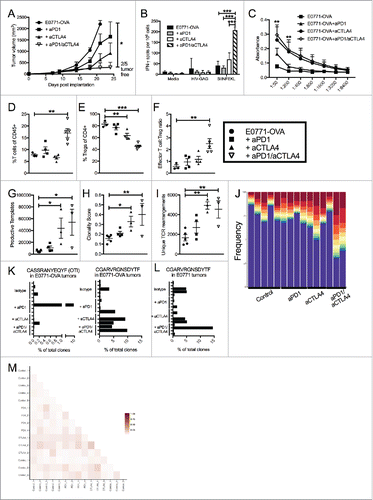
Figure 4. Targeting CTLA4 depletes intratumoral Tregs and enhances anti-tumor responses. (A) Flow cytometry intracellular staining of CTLA on CD4+ FoxP3- or CD4+ FoxP3+ T cells from the spleens of E0771-OVA tumor bearing mice and E0771-OVA tumors when tumors had reached a terminal endpoint. Representative histograms and summary data shown. Gray histogram represents staining control. (B) E0771-OVA cells were grown subcutaneously in C57 BL/6 mice and tumor volume was measured biweekly. Mice were randomized into groups and treated with anti-CTLA4 antibodies biweekly beginning when tumors measured >100 mmCitation3 or left untreated. (C) CD45+CD11b-CD4+FoxP3+ Tregs in tumors from mice in (B) at the terminal endpoint were quantified by flow cytometry. (D) Splenocytes from mice in (B) at the terminal endpoint were stimulated as indicated and IFN-γ producing cells were analyzed by ELISPOT. A-D representative of 3 experiments. Error bars indicate SEM. n = 5 per group for A-E; *P < 0.05; **P < 0.01; ***P < 0.001 by one-way ANOVA with Bonferroni's multiple comparisons test to E0771-OVA control group.
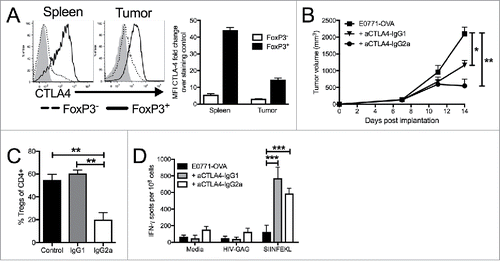
Figure 5. Manipulation of PDL1/PD1 enhances tumor targeting by immune cells. (A) ELISPOT assay for IFN-γ producing cells with OTI T cells as responders and E0771 parental, E0771-OVA or E0771-OVA PDL1 OE tumor cells as stimulators. (B) E0771 cell lines expressing luciferase were incubated with indicated ratios of OTI T cells and % of live cells as measured by luciferase expression compared to no OTI control is shown. (C) E0771-OVA or E0771-OVA PDL1-OE tumors were grown subcutaneously in C57 BL/6 mice and measured biweekly. (D) E0771-OVA GFP-CRISPR control or E0771-OVA PDL1 KO tumors were grown subcutaneously in C57 BL/6 mice and measured biweekly. (E) E0771-OVA GFP-CRISPR control or E0771-OVA PDL1 KO tumors were grown as before and on day 3 post tumor implantation, 1 × 106 OTI T cells were transferred IV into a cohort of each group. (F) Serum from terminal bleed of mice in (E) was analyzed by ELISA for anti-OVA antibodies. (G) Splenocytes from mice in (E) at the terminal endpoint were stimulated as indicated and IFN-γ production was analyzed by ELISPOT. A-B representative of 3 experiments. C-G representative of 2 experiments. Errors bars indicate SEM. n = 5 per group for A-F; *P < 0.05; **P < 0.01; ***P < 0.001 by 2-tailed Student's t test or one-way ANOVA with Bonferroni's multiple comparisons test to E0771-OVA control group.
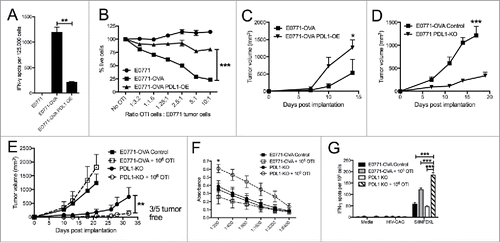
Figure 6. Unique patterns seen in mice that respond to treatment, regardless of what treatment. (A) E0771-OVA cells were grown subcutaneously in C57 BL/6 mice. Mice were randomized into groups and treated with anti-PD1 and anti-CTLA4-IgG2 a antibodies biweekly beginning when tumors measured >100 mmCitation3 or left untreated. Splenocytes from these mice were stimulated as indicated and IFN-γ production was analyzed by ELISPOT. (B) Serum from mice treated as in (A) was analyzed by ELISA for anti-OVA antibodies. Data shown in (A) and (B) is pooled from 2 independent experiments. (C) Correlation of tumor volume with the % of TILs. (D) Correlation of tumor volume with the % of Tregs. (E-H) Regrouping of data presented in . (E) Number of total productive TCR templates present in tumors. (F) Clonality score of TCRs present in tumors. (G) Number of unique TCR rearrangements present in tumors. (H) Frequency of the top ten TCR clones found in individual tumor samples. Each color (red through green) at the top of the bars represents the top 10 individual clones. The blue bar represents all remaining clones present in the sample. Error bars indicate SEM. Data are grouped as responders (tumors decreasing in size post treatment) and untreated/non-responders (tumors continuing to grow post treatment or left untreated). n = 6-9 per group for A and B; n = 3-5 per group for C-H; * P < 0.05; ** P < 0.01; *** P < 0.001 by two-tailed Student's t-test.
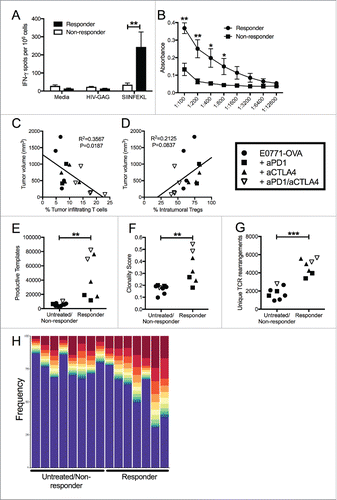
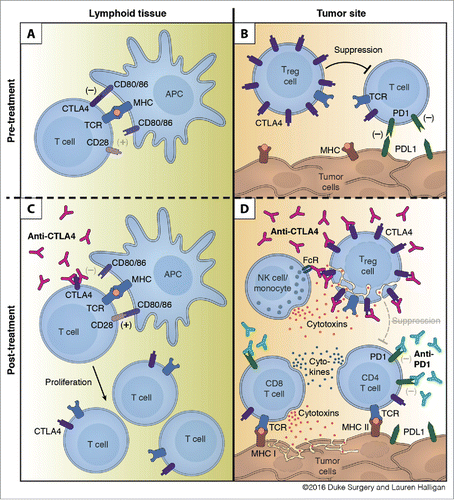
Figure 7. Mechanism of action for dual checkpoint blockade. (A) T cell expansion in the lymphoid tissue is hampered by negative signals from CTLA4. (B) Intratumoral Tregs and PDL1 expression by tumor cells dampen T cell responses within the tumor. (C) Treatment with anti-CTLA4 blocks engagement with CD80/86 and allows for expansion of T cells. (D) Binding of anti-CTLA4 to Tregs allows for blockade of contact mediated suppression of T cell responses and antibody-dependent cell-mediated cytotoxicity (ADCC) of Tregs by NK cells and/or monocytes. Anti-PD1 blocks engagement of PD1 with tumoral PDL1 and leads to enhanced cytokine production by CD4 and CD8 T cells and increased tumor cell killing by CD8 T cells.