Figures & data
Figure 1. Characteristics and validation of mouse genotypes. A, Breeding scheme. Nomenclature and abbreviations are presented in Table S1. B, Kaplan-Meier survival curves. PPARγ-deficiency in myeloid cells such as macrophages (MPh/MPH) but not in the intestinal epithelium (IEC) shortens the life span compared with WT mice (*p < 0.0001 RAS PPARΔMPH (n = 53) vs. WT (n = 149), log-rank test). Legend: “1” = event (death); “0” = alive or censored. C, Western blots of pull-down assays detecting active GTP-bound pan-RAS proteins in mouse whole tissue lysates compared to total pan-RAS (input) using RALGDS-GST as a bait. Representative gels are presented. KRASG12V-mutant human CRC cell line SW480 was a positive control. D, Pparg exon1/2 excision. Genomic PCRs were conducted on DNAs isolated from organs of the respective genotypes. Representative agarose gels are presented. Expected amplicon sizes are indicated.Citation49 E, PPARγ protein expression. FFPE tissue sections from WT and PPARΔIEC mice colon (Co) were stained with PPARγ Ab for immunohistochemistry (IHC). Magnification 200x. F, PPARγ target gene expression. Total RNA was extracted from snap-frozen small intestine (SI). CT-values from RT-qPCRs on acyl-CoA oxidase (Aco) were normalized to β2-microglobulin (B2 m) and calculated as -fold ± S.E. (n = 3 per genotype, *p < 0.05, Mann Whitney test). G, PPARγ and RAS target protein expression. Whole tissue lysates (from Co and SI) were subjected to Western blotting. Representative gels and quantitative analysis are shown. O.D. values of bands in gels were normalized to HSP90 and calculated as -fold ± S.E. compared with WT mice (n = 2 per genotype, *p < 0.05, Wilcoxon signed rank test).
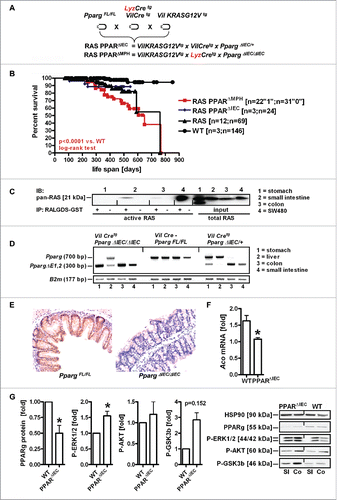
Figure 2. PPARγ-deficiency exacerbates pre-malignant phenotypes driven by mutant KRAS in the mouse small intestine. A, Exemplary microscopic images of H&E staining in FFPE tissue sections from RAS and PPARΔIEC mice. Multiplicity and incidence of histopathological parameters in mouse genotypes are presented in Table S2. Top left: Hyperplastic epithelium with goblet cells in the lower small intestine (SI); Top right: Tubular adenoma in the duodenum; Bottom left: hyperplastic polyp (HPP); Bottom right: serrated adenoma (SA) with dysplasia. Magnification 100x and 200x. B, Quantitative analysis of H&E histopathology in A. Left: As a surrogate indicator for transmural inflammation, the distance/diameter (length) between the crypt base and the tunica adventitia (serosa) were measured (in μm). Data are means ± S.E. (n≥4 mice, *p < 0.05, Mann Whitney test). Middle: Number of cases (mice) having more or less than 5 polyps per total SI (*p < 0.05, Fisher Exact test). Right: Mice having discrete or extended polyps per total SI (*p < 0.05, Fisher Exact test). C, Mutant KRAS and PPARγ-deficiency increase immune cell numbers in the lamina propria of the non-malignant mucosa compared with WT mice. FFPE tissue sections of WT, RAS and RAS PPARΔIEC mice were stained using IHC with Abs detecting B-cells (B220/CD45R), T-cells (CD3), Tregs (FOXP3) and macrophages (F4/80). Staining intensity and frequency were scored. Data are means ± S.E. (n≥3 per genotype, *p < 0.05, Kruskal Wallis test).
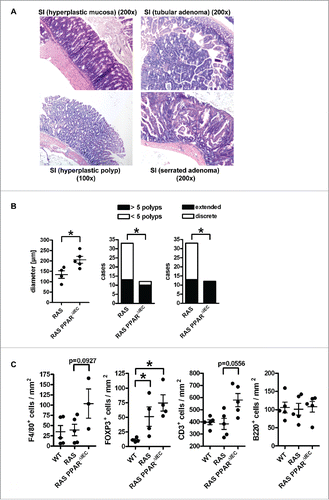
Figure 3. Mutant KRAS promotes senescence and immune escape in serrated polyps independently of PPARγ. A, IHC on tissue sections of RAS and RAS PPARΔIEC mice (n≥3 per genotype) with Abs detecting P-ERK1/2 and P21 as a marker for oncogene-induced senescence (OIS). Left: P-ERK1/2 positivity was reduced in polyps (arrow “p”) compared to the strong signal in crypts and basal regions of villi in the adjacent mucosa (arrow “m”); Right: P21 was increased in apical tips of polyps and interspersed lamina propria cells. No difference was observed between polyps of the two genotypes (not shown). B, Quantitative analysis of IHC in A. Number of mice having high (scores +2,+3) or low (scores 0,+1) positivity in polyps vs. non-malignant mucosa (*p< 0.05, Fisher Exact test). C, IHC using Abs against B-cells (B220/CD45R), T-cells (CD3), Tregs (FOXP3) and macrophages (F4/80) was calculated as in B (*p< 0.05, Fisher Exact test). Macrophages remained high in polyps, T-cells were reduced.
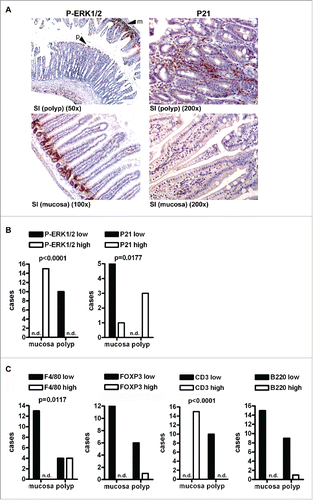
Figure 4. RAS-mediated polyp load and proliferation are reduced by PPARγ-agonist. A, Microscopic images of H&E staining. Left: Swiss roll showing the complete SI from a chow-fed RAS mouse with extended serrated polyps; Right: rosi-fed RAS mouse. Magnification 10x. B, Quantitative analysis of H&E histopathology. Left: Transmural inflammation. Data are means ± S.E. (n≥6 mice per group, n.s., Mann Whitney test). Middle: Mice with more or less than 4 polyps per SI (n.s., Fisher Exact test). Right: Mice having discrete or extended polyps per SI (p = 0.0505, Fisher Exact test). C, Quantitative analysis of body and spleen weights. Data are means ± S.E. (n≥10 mice per treatment group and genotype, *p < 0.05 vs. ROSI, One-way ANOVA). D, IHC on tissue sections of RAS and RAS+ROSI mice with an Ab detecting the proliferation marker Ki67 in the non-malignant mucosa. Data are means ± S.E. (n≥4 per group, *p < 0.05 vs. ROSI, Kruskal Wallis test). Representative images are shown in A.
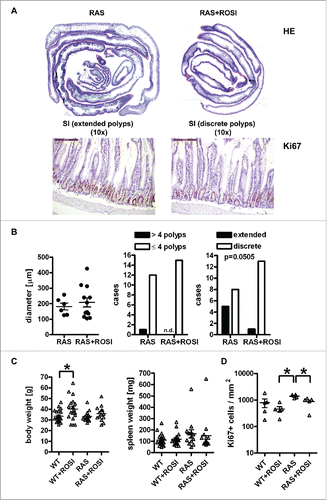
Figure 5. RAS-mediated phenotypes are attenuated by PPARγ-agonist. A, Immune marker gene expression in mouse tissues. Total RNA was extracted from snap-frozen SI of WT and RAS mice. CT-values from RT-qPCRs normalized to B2 m were calculated as -fold ± S.E. (n = 6 per genotype, *p < 0.05 WT vs. RAS for all genes, Mann Whitney test). B, IHC on tissue sections of RAS and RAS+ROSI mice using Abs detecting T/B cells and total macrophages in the non-malignant mucosa. Data are means ± S.E. (n = 3 per group, p = 0.100, Mann Whitney test). C, PPARγ-activation increases intestinal M1 macrophage numbers. IHC with Abs detecting subpopulations of macrophages. Data are presented as in B (n≥4 per group, *p < 0.05 vs. ROSI, Mann Whitney test).
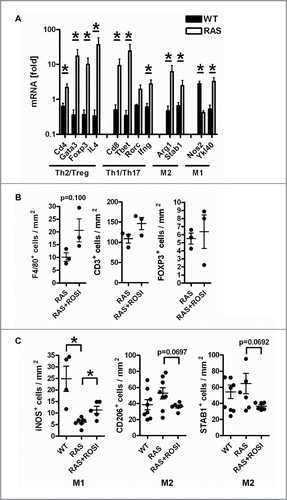
Figure 6. Immune-related marker genes/proteins are differentially regulated by PPARγ-agonist. A, IHC on tissue sections of RAS and RAS+ROSI mice using Abs detecting subpopulations of T-cells (top) and macrophages (bottom) in polyps vs. the non-malignant mucosa. Data are means ± S.E. (n≥3 per group and genotype, *p < 0.05 vs. ROSI, Mann Whitney test). B, Total RNA was extracted from snap-frozen SI of RAS and RAS+ROSI mice. CT-values from RT-qPCRs normalized to B2 m were calculated as -fold ± S.E. (n = 9-10 per group, Mann Whitney test). Il4 (*p < 0.05) was reduced by rosi; there was a trend for Cxcl10 (p = 0.08) up-regulation.
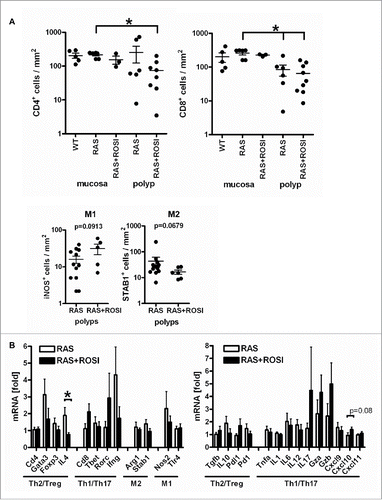
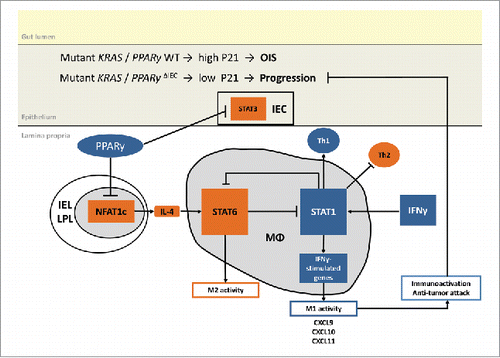
Figure 7. Proposed model of RAS and PPARγ crosstalk. Suggested mechanism based on published literature and own experimental data: PPARγ-activation reshapes the immune cell composition in the lamina propria from an immuno-suppressive towards a tumor-attacking profile. For example, PPARγ may reduce NFAT-driven Il4 (Th2/M2) gene expression to indirectly activate “killer” type responses (Th1/M1).