Figures & data
Figure 1. Pulmonary PGIS overexpression inhibits CMT167 tumor progression and metastasis. CMT167 tumors were implanted in pulmonary PGIS overexpressing transgenic mice (PGIS OE, n = 15) or littermate controls (WT, n = 21). After four weeks, lungs and livers were harvested and imaged ex vivo with an IVIS 50 imaging system. (A) Primary lung tumor volumes were measured; each point represents a single tumor; ** = p < 0.01. (B) The incidence of secondary pulmonary tumors and liver metastases was quantified by ex vivo bioluminescence (percent of total mice per group; * = p < 0.05). (C) Representative ex vivo bioluminescent images of livers with metastases from a control mouse (left panel) and a PGIS overexpressor (right panel). The number (D) and signal intensity (E) of pulmonary and liver metastases were quantified by ex vivo bioluminescence imaging.
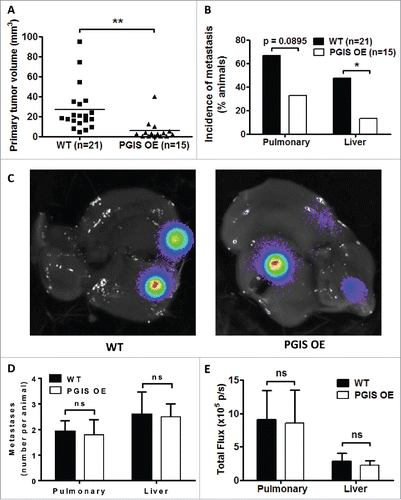
Figure 2. Pulmonary PGIS overexpression increases lymphocyte infiltration in CMT167 tumors. Representative CD3 (top panels), CD4 (middle panels), and CD8 (bottom panels) immunostaining (AF488, green) of sections from PGIS overexpressing mice (right panels) and wildtype littermates (left panels). Nuclei were stained with DAPI (blue); scale bar = 40 μm.
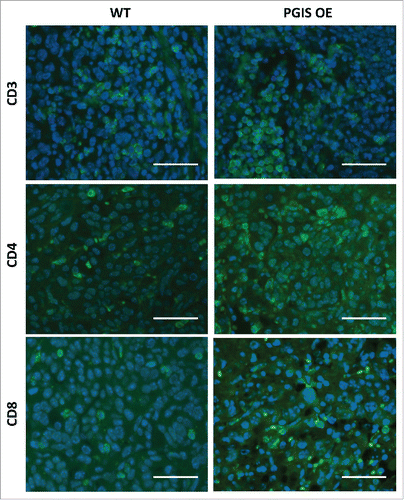
Figure 3. Pulmonary PGIS overexpression increases CD4+ T cell infiltration in CMT167 tumors. Three sections from each animal were stained for (A) CD3+, (B) CD4+, and (C) CD8+ TILs and three random fields (20x) per section were quantified. The mean was used for analysis; each point represents a single mouse. (D) Mice were injected with either anti-CD4 monoclonal antibody or control IgG antibody starting 1 day prior to tumor implantation and throughout the course of the tumor progression experiment. CMT167 tumors were implanted in either PGIS OE (n = 5) or WT littermates (n = 4). After four weeks, primary lung tumors were isolated and tumor volumes were measured by digital calipers. Each point represents a single tumor. (E) Two weeks after CMT167 tumor implantation, total RNA was isolated from tumor-bearing left lungs. CXCL9 and CXCL10 mRNA expression was analyzed by quantitative RT-PCR and normalized to β-actin. Results represent the mean with the S.E.M. indicated. Statistically significant differences are indicated as calculated by unpaired t test, n.s. = not significant.
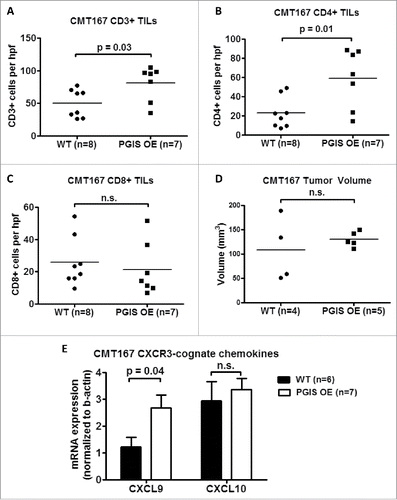
Figure 4. Pulmonary PGIS overexpression has no effect on LLC orthotopic lung tumor progression and metastasis. LLC tumors were implanted in pulmonary PGIS overexpressing transgenic mice (PGIS OE, n = 5) or littermate controls (WT, n = 5). After 3 weeks, lungs, livers, and brains were harvested and imaged ex vivo with an IVIS 50 imaging system. (A) Primary tumor volumes were measured by digital calipers. Each point represents a single tumor. The incidence (B), number (C) and signal intensity (D) of liver and brain metastases were quantified by ex vivo bioluminescent imaging, ns = not significant.
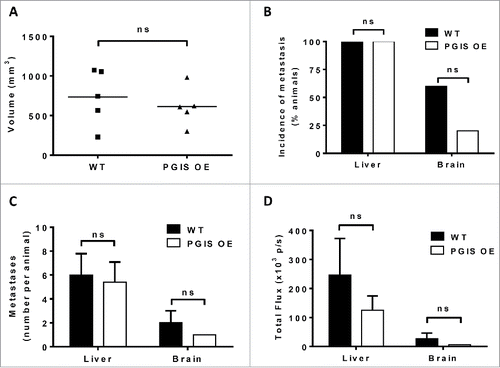
Figure 5. Pulmonary PGIS overexpression has no effect on lymphocyte infiltration in LLCtumors. Representative CD3 (top panels), CD4 (middle panels), and CD8 (bottom panels) immunostaining (FITC, green) of sections from PGIS overexpressing mice (right panels) and wildtype littermates (left panels). Nuclei were stained with DAPI (blue); scale bar = 100 μm.
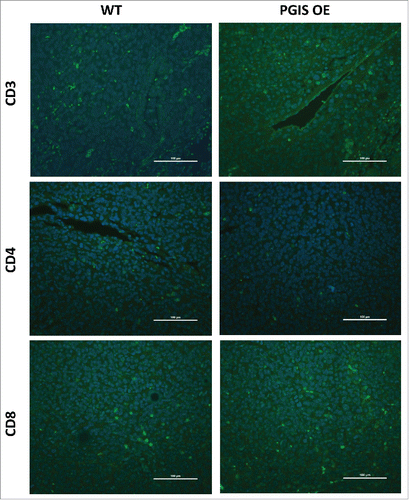
Figure 6. Pulmonary PGIS overexpression has no effect on LLC tumor-infiltrating lymphocytes. Three sections from each animal were stained for (A) CD3+, (B) CD4+, and (C) CD8+ TILs, and three random fields (20x) per section were quantified. The mean was used for analysis; each point represents a single mouse. (D) Two weeks after LLC tumor implantation, total RNA was isolated from tumor-bearing left lungs. CXCL9 and CXCL10 mRNA expression was analyzed by quantitative RT-PCR and normalized to β-actin. Results represent the mean with the S.E.M. indicated. Differences between groups were compared using unpaired t test, n.s. = not significant.
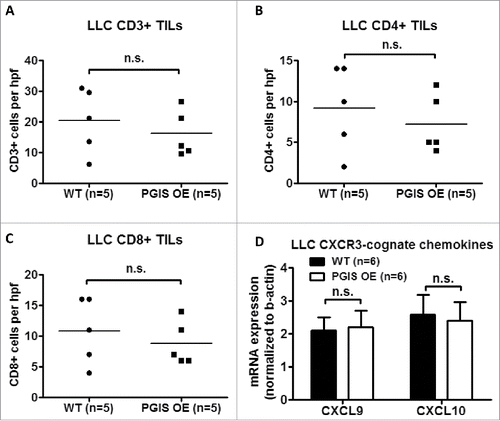
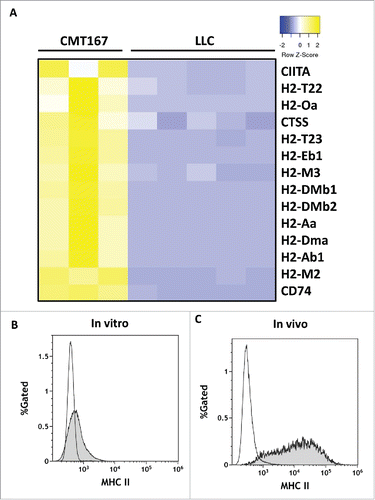
Figure 7. In vivo, CMT167 cells but not LLC cells express MHC class II genes and cofactors necessary for MHC class II processing and presentation. CMT167 tumors and LLC tumors were implanted in GFP-expressing transgenic mice. (A) After several weeks, cancer cells were recovered by sorting for GFP-negative cells using flow cytometry. Total RNA was isolated, analyzed by RNA-seq, and compared with total RNA collected from cancer cells grown in vitro. CMT167 and LLC cancer cells were analyzed for surface expression of MHC II by flow cytometry. The histograms show overlays of MHC II expressed by CMT167 cells (shaded) and LLC cells (white) in vitro (B) and in vivo (C). Data are representative of six independent in vitro experiments and three independent in vivo experiments (n = 2 mice per group).