Figures & data
Figure 1. Expression of LLT1 and CD161 in NSCLC tumors. (A-H) Hematoxylin (HE) counterstained IHC stainings of (A-D) FFPE and (E-H) frozen sections of two representative tumors from untreated NSCLC patients using (A-D) anti-human LLT1 clone 2F1 and (E-H) anti-human CD161 clone DX12. (B-D) represent higher magnifications (x100) of areas (black rectangles) in (A) (magnification x10). (F-H) represent higher magnifications (x100) of areas (black rectangles) in (E) (magnification x10). (A-D) Solid adenocarcinoma (ADC) subtype. (E-H) Lepidic ADC subtype. Str, Stroma; Tu, Tumor Nests.
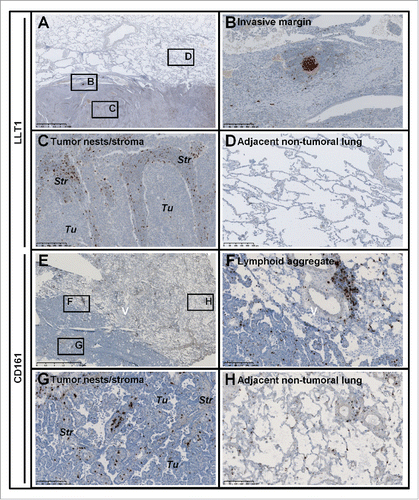
Figure 2. LLT1co-localizes with GC-B cells within NSCLC-associated TLS and with CD3+ (NKp46−) T cells within tumor stroma. (A) HE counterstained IHC staining of a FFPE section of tumor from untreated NSCLC patient using anti-human LLT1 clone 2F1, with a focus on a TLS at high magnification (x100). (B,C) represent the same TLS on serial sections stained with anti-human CD20 and anti-human Ki67 (B) or anti-human CD20 and anti-human CD21 (C). (D) Correlation between the number of LLT1+ follicles and the number of CD21+ follicles in 32 tumors from untreated NSCLC patients. Linear regression curve, spearman r value and p value (95% confidence interval) are shown. (E-K) Double IF stainings of FFPE sections from NSCLC tumors (magnification x200) using anti-human LLT1 clone 2F1 associated with anti-human CD20 (E, G), anti-human CD3 (F, H), anti-human CD19 (J), or anti-human NKp46 (K). (E, F) represent the same TLS on serial sections. (G-J) represent the same area of tumor stroma on serial sections. K represents another area of NSCLC tumor stroma.
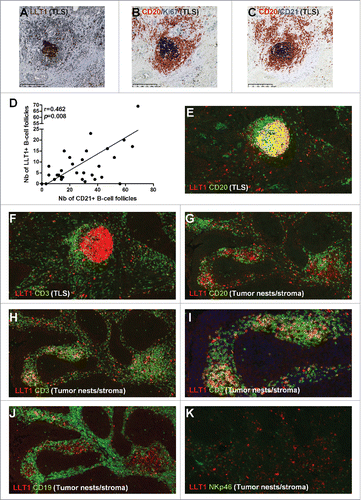
Figure 3. LLT1 is expressed on GC-B cells in NSCLC tumors. (A) Representative dot plots of LLT1 expression by flow cytometry on all B cells and B cell subsets in two different NSCLC tumors (upper and lower panels), using anti-human LLT1 mAb clone 4F68 and isotype control. Percentages of cells expressing LLT1 among each B cell subset in NSCLC tumors are indicated, n = 10. (B) Representative dot plots of LLT1 expression on GC/pre-GC B cells in a NSCLC tumor and matched non-tumoral distant lung, with percentages of cells expressing LLT1 among GC/pre-GC B cells in each compartment, n = 10. (C) Representative dot plots of LLT1 expression on different cell subsets in a NSCLC tumor, with percentages of cells expressing LLT1 among each cell subset in NSCLC tumors, n = 10. Mean percentages are indicated by red horizontal lines. P values were calculated using (A, C), One-way Anova, Kruskal-Wallis test, Dunn's Multiple Comparison Test and (B), unpaired t test. *p<0.05, **p<0.005.
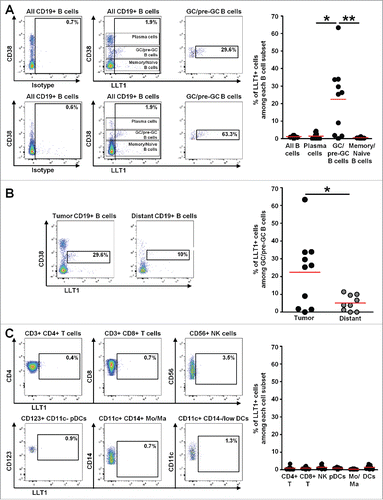
Figure 4. Accumulation of CD161-expressing CD3+ T cells within NSCLC tumor stroma and at the vicinity of TLS. (A-B) IF staining of a frozen section from NSCLC tumor using anti-human CD161 clone DX12 (B) or isotype control (A) associated with DAPI. (C-E, H) Double IF stainings of serial frozen sections from the same NSCLC tumor using anti-human CD161 clone DX12 associated with (C) anti-human CD20, (D-E, H) anti-human CD3, and (H) DAPI. (E) represents a higher magnification (x400) of area (white rectangle) in (D). (F) Double IF staining of a serial frozen section from the same NSCLC tumor using anti-human LLT1 clone 2F1 associated with anti-human CD20. (G) ImageJ-processed overlay of LLT1 IF staining from (F) and CD161/CD20 double IF staining from (C), at high magnification. (I-J) Triple IF stainings of frozen sections from two NSCLC tumors using anti-human CD161 clone DX12 associated with anti-human CD20 and anti-human CD3. (A-G, I-J) focus on TLS. (H) focus on tumor stroma and tumor nests. (A-D, F, H-J) magnification x200. (E, G) magnification x400.
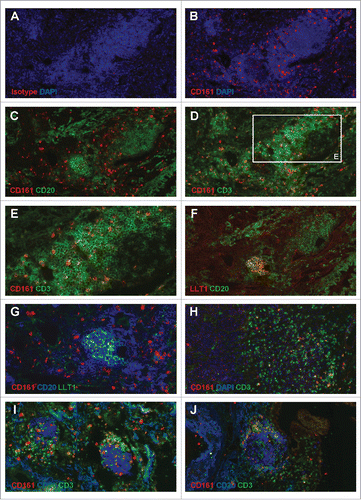
Figure 5. High frequencies of CD161-expressing CD4+ T cells in NSCLC tumors. (A) Histogram representing the mean percentages of each cell subset among viable CD45+ CD161+ lymphocytes, in each compartment. (B) Histogram representing the percentages of cells expressing CD161 among each cell subset, in each compartment, n = 33. Means are indicated by red horizontal lines. P values were calculated using One-way anova/Kruskal-Wallis/Dunn's test. (C) Graphs representing the percentages of cells expressing CD161 among each cell subset, in each compartment, in matched patients, n = 24. P values were calculated with One-way anova/Friedman/Dunn's test. *p<0.05, **p<0.005, ***p<0.001.
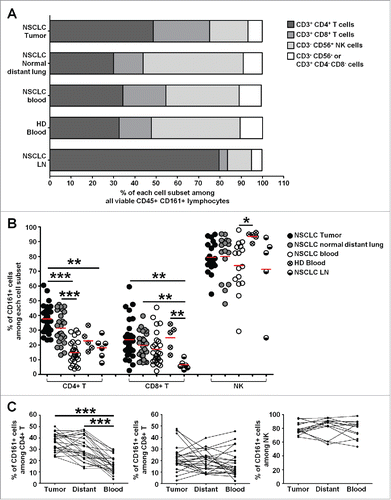
Figure 6. Heterogeneity of CD161-expressing CD4+ and CD8+ T cells within NSCLC tumors. (A-B) Correlation factors between KLRB1 gene expression and expression of genes from the “PanCancer Immune Profiling Panel” (NanoString Technologies) in highly purified intra-tumoral CD4+ T cells (A) or CD8+ T cells (B). Genes significantly correlated with KLRB1 (FDR < 0.05, 0.05 < FDR < 0.10, and up to 0.10 < FDR < 0.15) are shown. Vertical dashed lines represent limits for correlation factors <-0.5 and >+0.5. (C-D) Potential associations between genes positively (C) and negatively (D) correlated with KLRB1 in CD4+ T cells (FDR < 0.15) with corresponding pathways determined using ClueGo software.
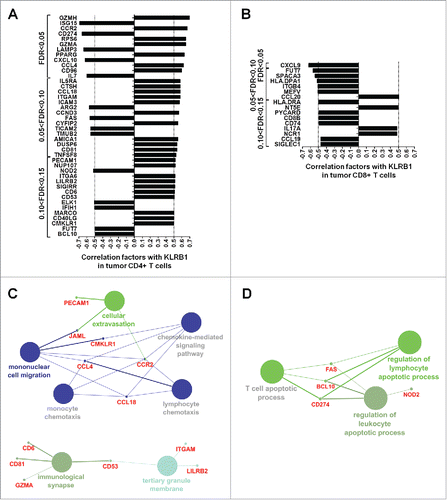
Figure 7. Higher frequencies of CD161-expressing central-memory CD4+ T cells in NSCLC tumors as compared to distant lung and peripheral blood. (A) Rings illustrating the mean percentages of naive, effector memory (EM), central memory (CM) and effectors (EMRA) subsets within CD161− CD4+ or CD161+ CD4+ T cells (left panels), or within CD161− CD8+ or CD161+ CD8+ T cells (right panels), in each compartment, n = 12. (B) Histograms representing the percentages of CD161 expression within each CD4+ (left panel) and CD8+ (right panel) T cell subsets in each compartment, n = 12. Means are indicated by red horizontal lines. *p <0.05 (One-way anova/Kruskal-Wallis/Dunn's test). (C) Mean percentages of CD161 expression within each CD4+ (left panel) and CD8+ (right panel) T cell subsets in matched compartments, n = 11. *p<0.05, **p<0.005 (One-way anova/Friedman/Dunn's test).
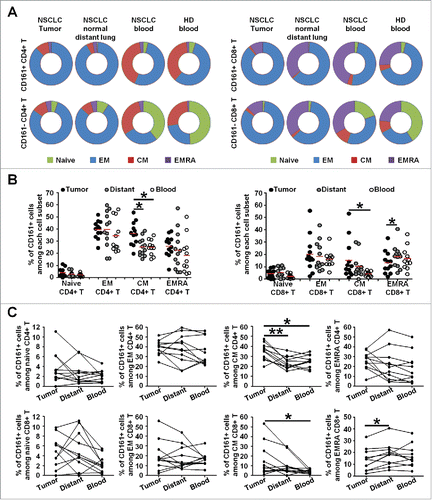
Figure 8. CD161-expressing CD4+ and CD8+ T cell phenotypes in NSCLC tumors. (A-B) Histograms represent the percentages of cells producing Granzyme B (GzB), IFN-γ or TFN-α among PMA/Ionomycin-stimulated CD161− or CD161+ CD4+ T cells (A), and CD161− or CD161+ CD8+ T cells (B) in NSCLC tumors, n = 18. (C-D) Upper heat maps illustrate the percentages of cells producing Granzyme B (GzB), IFN-γ or TFN-α among PMA/Ionomycin-stimulated CD4+ CD161− T cells and CD4+ CD161+ T cells (A) or CD8+ CD161− T cells and CD8+ CD161+ T cells (B) in NSCLC tumors, n = 18. Lower heat maps illustrate the percentages of cells expressing CD38 (n = 14), CD69 (n = 11), CD96 (n = 9), CD30L (n = 9), CD40L (n = 11), OX40 (n = 11), 4-1BB (n = 11), CD27 (n = 13), PD-1 (n = 15), Tim-3 (n = 13) or CD103 (n = 12) among CD4+ CD161− T cells and CD4+ CD161+ T cells (A), or CD8+ CD161− T cells and CD8+ CD161+ T cells (B) in NSCLC tumors. P values were calculated using Wilcoxon (paired, non-gaussian) test between CD161− and CD161+ cells. *p<0.05, **p<0.005, ***p<0.001.
Figure 9. Correlation of CLEC2D and KLRB1 with clinical outcome. (A-B) Forest plot depicting the meta-analysis of (A) CLEC2D and (B) KLRB1 gene expression associated with better clinical outcome in different cohorts of NSCLC patients as indicated. Global p values are indicated. The width of the diamond shows the confidence intervals for each Hazard ratio. HR, Hazard Ratio. (C-D) Kaplan-Meier curves of overall survival among the GSE50081 cohort, according to CLEC2D gene expression combined with estimated B cell density (C), or KLRB1 gene expression combined with estimated cytotoxic cell density (D), after transformation of each variable into binary (high or low) variables (median cut-off value).
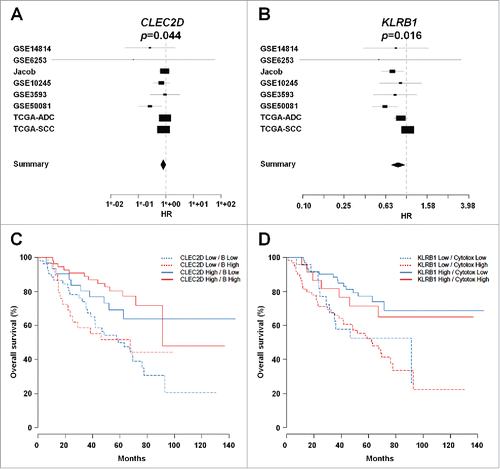
Table 1. Clinical and pathological features of the 33 untreated patients with NSCLC used in flow cytometry analysis.
Table 2. Clinical and pathological features of the 31 untreated patients with NSCLC used in gene expression analysis.
