Figures & data
Figure 1. Heatmap displaying a differential gene expression in a color scale in HLA class I-mutant gliomas vs. wild-type HLA class I gliomas. Tumors harboring HLA class I mutations overexpress lymphocyte killer effector genes. As previously defined, “Cytolytic activity” is included as a single gene and determined by Granzyme A (GZMA) and Perforin (PRF1), two cytolytic genes expressed in activated CD8+ T-cells.Citation60 *P < 0.05 represents the significance of the association between the expression of each gene and HLA class I mutant-gliomas. [Adapted by permission from Springer Nature: Nature Biotechnology, Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes, Shukla et al., copyright 2015]Citation59.
![Figure 1. Heatmap displaying a differential gene expression in a color scale in HLA class I-mutant gliomas vs. wild-type HLA class I gliomas. Tumors harboring HLA class I mutations overexpress lymphocyte killer effector genes. As previously defined, “Cytolytic activity” is included as a single gene and determined by Granzyme A (GZMA) and Perforin (PRF1), two cytolytic genes expressed in activated CD8+ T-cells.Citation60 *P < 0.05 represents the significance of the association between the expression of each gene and HLA class I mutant-gliomas. [Adapted by permission from Springer Nature: Nature Biotechnology, Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes, Shukla et al., copyright 2015]Citation59.](/cms/asset/2de7f07e-d6a3-4342-85f3-3691c7537d56/koni_a_1445458_f0001_oc.jpg)
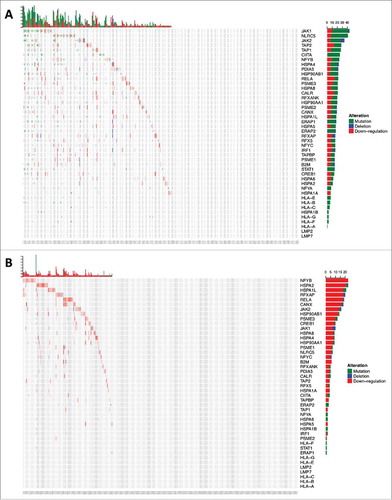
Figure 2b. Cumulative HLA class I and APM defects across gliomas from TCGA. (A) 213/396 GBM and (B) 166/511 LGG samples were found to have alterations in the formation of the HLA class I peptide complex that enable glioma cells to escape immune recognition by CD8+ T-cells. (C) Highlighted in red are the components showing different types of alterations in antigen processing and presentation ranging from defects in the IFN-γ pathway (JAK1, JAK2, STAT1) and the HLA class I enhanceosome (NLRC5, CIITA, IRF1, RFX5, RFXANK, RFXAP, X2BP, NF-Y) that mediate the transcriptional activation of HLA class I genes; PSME1, PSME2, PSME3 genes coding for the alpha, beta, and gamma subunits that make up the proteasome activator complex PA28 for peptide generation; ERAP1 and ERAP2 that trim longer precursor of antigenic peptides; HSPA and HSPC that chaperones the peptides to their loading on the MHC class I in the endoplasmic reticulum; and TAP1, TAP2, TAPBP, calreticulin, calnexin, PDIA3, β2-microglobulin and HLA class I genes that participate in the formation of the MHC class I peptide complex.
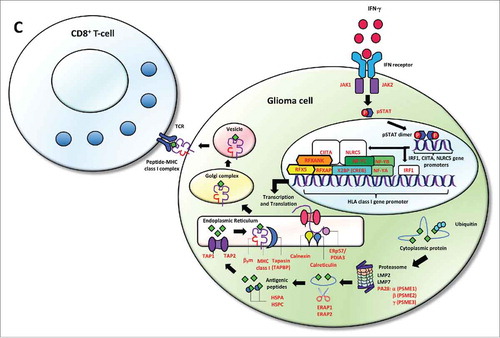
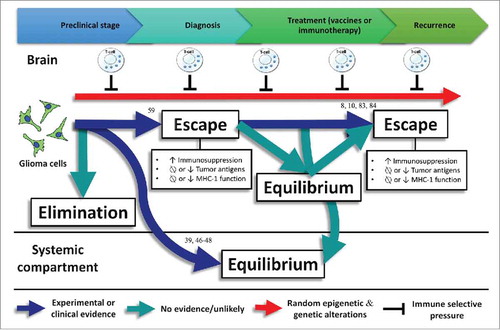
Figure 3. Experimental or clinical evidence regarding immune editing in gliomas with numbers representing the references related to this evidence. No evidence exists about the elimination phase in gliomas. Commonly, gliomas do not spread outside the central nervous system. However, glioma metastases have been detected in transplanted organs of immunosuppressed transplant recipients, suggesting that the equilibrium phase is taking place in the systemic compartmentCitation39,Citation46-Citation48 Associations between loss-of-function HLA class I mutations and upregulation of lymphocyte killer effector genes,Citation59 as well as HLA class I and APM defects have been found in gliomas, evidencing the escape phase. In addition, loss of glioma-specific antigens after using targeted immunotherapies lead to a negative selection of tumor variants cells expressing these antigens.Citation8,Citation10,Citation83,Citation84