Figures & data
Figure 1. Gene expression presented in Transcripts per million (TPM) according to disease progression status for TOX (A), FYB (B), and CD52 (C) in all CTCL samples. Quartile boxplots are shown.
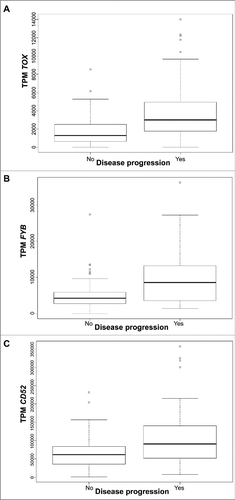
Figure 2. Survival analysis plots of all CTCL samples according to gene expression for TOX (A), FYB (B), and CD52 (C). Samples are dichotomized based on gene expression levels in TPM after finding an optimal cut-off; samples with TPM below cut-off labelled as low (solid line) and samples with TPM above cut-off labelled as high (dotted line). Survival plots correspond to Cox proportional hazards regression models.
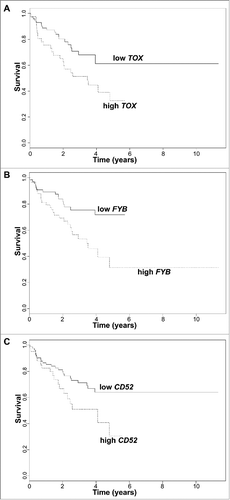
Figure 3. Gene expression presented in Transcripts per million (TPM) according to disease progression status for TOX (A), FYB (B), CD52 (C), and CCR4 (D), in early stage (≤IIA) CTCL samples. Quartile boxplots are shown.
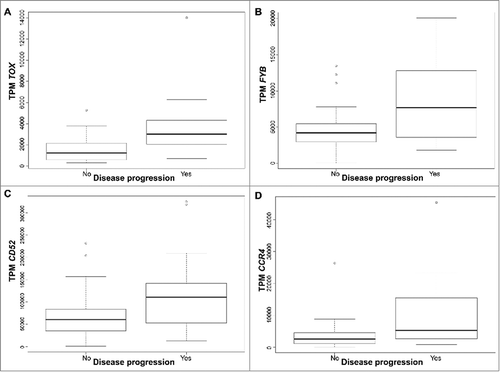
Table 1A. External validation of CTCL progression-associated genes in two pooled independent cohorts of Sézary patients with RNA-Sequencing gene expression data (Ungewickell et al., Choi et al.).
Table 1B. External validation of CTCL progression-associated genes in the Wang et al. independent cohort of Sézary patients with RNA-Sequencing gene expression data.
Figure 4. (A) Neutrophil immune cell fraction obtained in silico using CIBERSORT according to disease progression status in early stage (≤IIA) CTCL samples. Quartile boxplots are shown. (B) Survival analysis plots of late stage (IV) CTCL samples according to NK T lymphocyte fraction. Samples are dichotomized according to NK T lymphocyte immune cell fraction obtained in silico using CIBERSORT; samples with fraction below 0.05 labelled as low (solid line) and samples with fraction above 0.05 labelled as high (dotted line). Survival plot corresponds to Cox proportional hazards regression models.
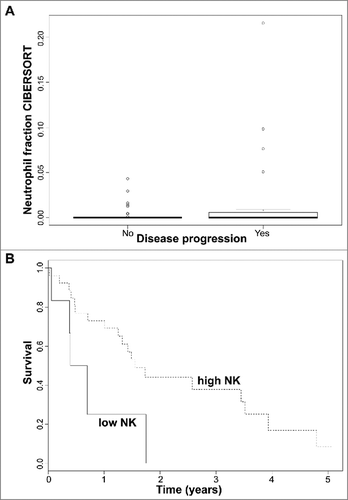
Table 2A. IL-17A through IL-17F expression in two pooled independent cohorts of Sézary patients with RNA-Sequencing gene expression data (Ungewickell et al., Choi et al.).
Table 2B. IL-17A through IL-17F expression in the Wang et al. independent cohort of Sézary patients with RNA-Sequencing gene expression data.
