Figures & data
Table 1. Summary of patient cohort information.
Figure 2. Hierarchical clustering dendrograms of the expression patterns of differently expressed microRNAs that can basically distinguish between high TMB and low TMB in lung adenocarcinoma.
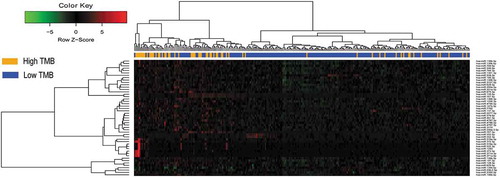
Figure 3. LASSO model and principal component analysis. (a) 10‑fold cross‑validation for tuning parameter selection in the LASSO model. (b) PCA prior to and (c) after LASSO variable reduction. LASSO, least absolute shrinkage and selection operator; PCA, principal component analysis.
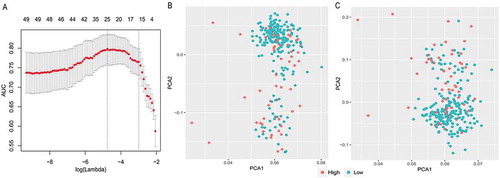
Table 2. Performance of 25-miRNA-based classifiers of tumor mutation burden in lung adenocarcinoma.
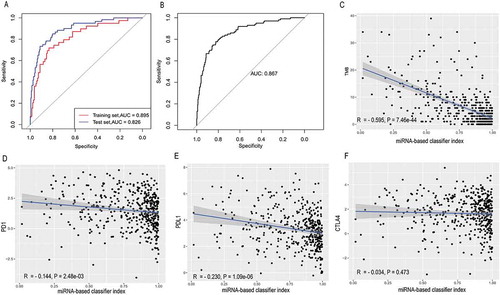
Figure 4. Receiver operating characteristic curves for the 25-miRNA-based signature index and its correlation with TMB, PD-1, PD-L1 and CTLA-4. (a) Receiver operating characteristic analyses in the training and the test set. (b) Receiver operating characteristic analyses in the total set. (c) The 25-miRNA-based signature index is highly correlated with TMB (d) The 25-miRNA-based signature index shows low correlation with PD-1 expression. (e) The 25-miRNA-based signature index shows low correlation with PD-L1 expression. (f) The 25-miRNA-based signature index is not correlated with CTLA4 expression.