Figures & data
Figure 1. FH changes the morphology and prolongs the viability of monocytes
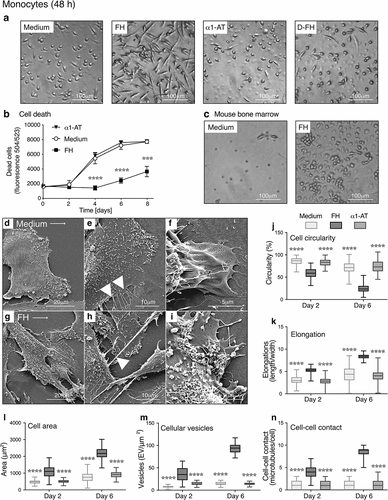
Figure 2. FH differentiates monocytes into macrophages
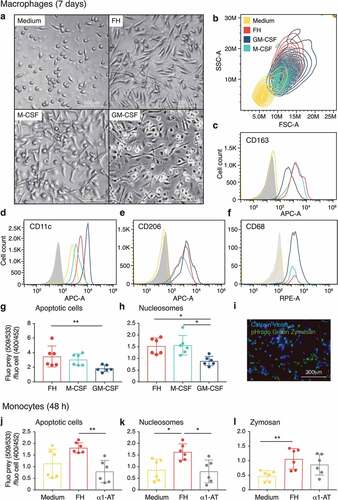
Figure 3. The morphological changes upon FH stimulation in CD14+ monocytes are induced by CCP19-20 and are inhibited by plasma
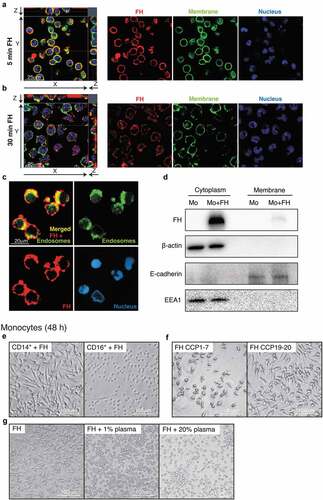
Figure 4. Transcriptome analysis of FH-stimulated monocytes
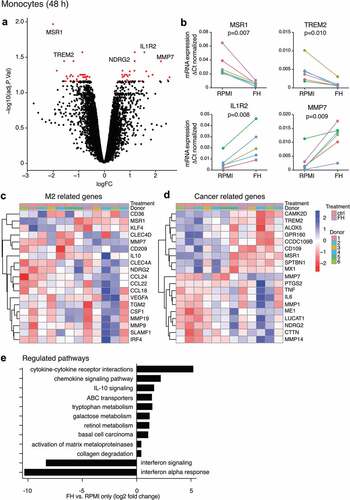
Figure 5. FH induces an anti-inflammatory cytokine release profile
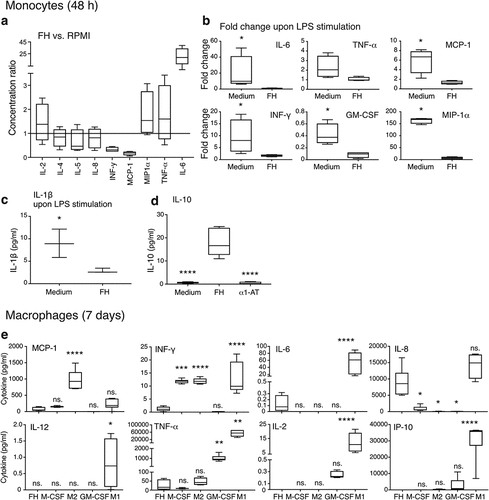
Figure 6. FH-induced macrophages exhibit a suppressive phenotype
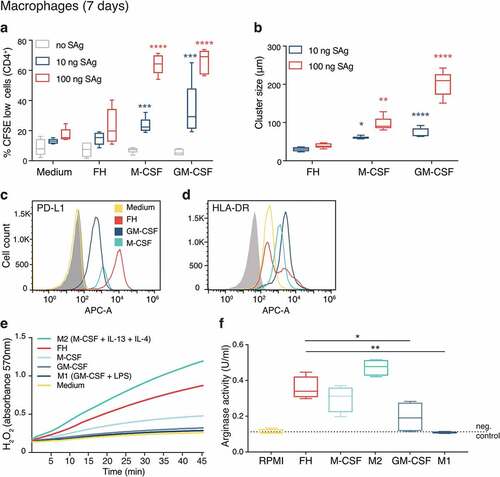
Figure 7. FH produced in breast tumors correlates with recurrence, disease severity and occurrence of M2 macrophages
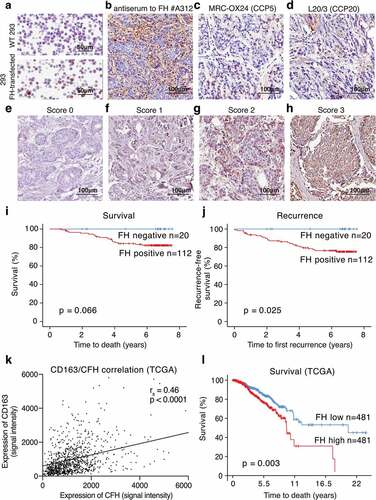
Table 1. Association between FH and clinical parameters
