Figures & data
Table 1. Proportion of MC38 IC-implanted mice.
Figure 1. Two profiles of CRC development in immunocompetent B6 mice. a, b.
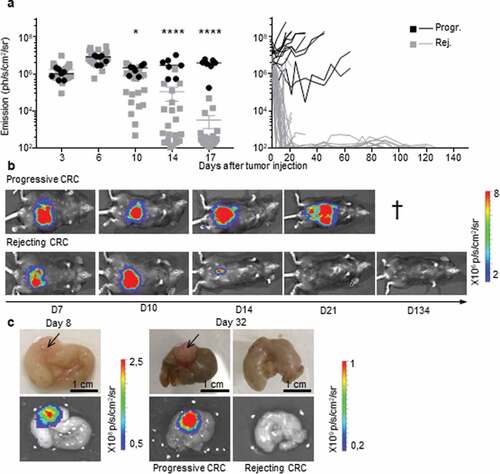
Figure 2. Rejection of MC38-fLuc tumors leads to elimination.
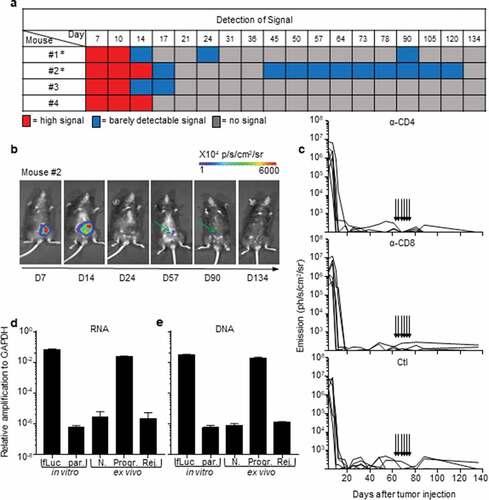
Figure 3. An immune-colon dependent effect generates the two CRC development profiles. a to c.
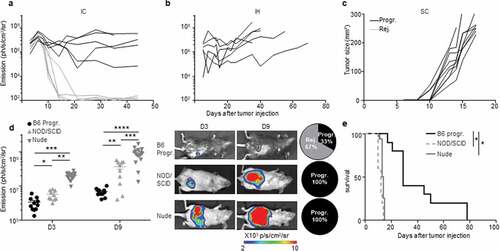
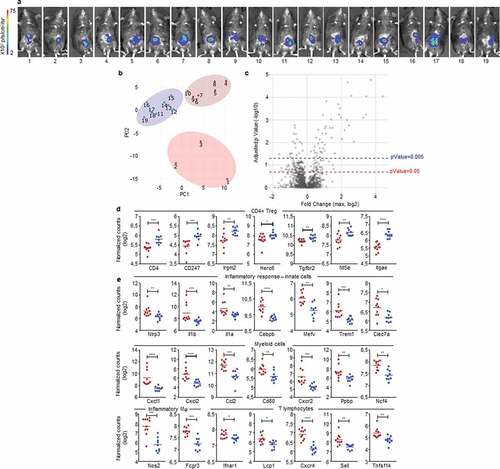
Figure 4. The immunosuppressive myeloid cell-related microenvironment is characterized in progressive CRC mice. a, b.
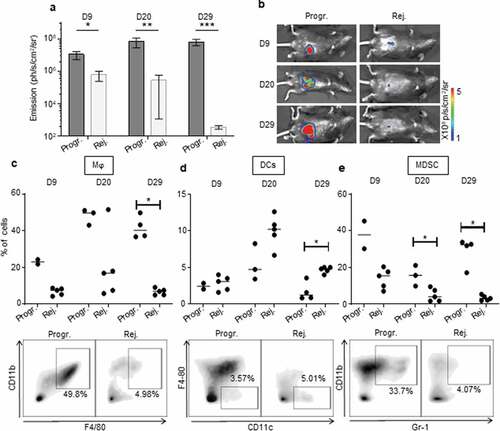
Figure 5. High CD8 T cells and low Treg infiltration in rejecting CRC tumors.
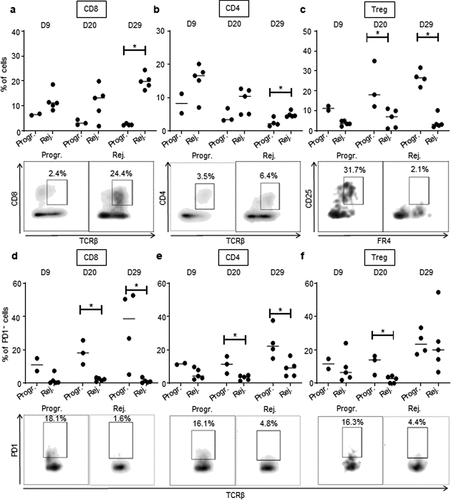
Figure 6. Early detection of opposing immune microenvironments in CRC tumors.