Figures & data
Figure 1. Loss of ATG7 blocks autophagy and sensitizes cells to nutrient deprivation.
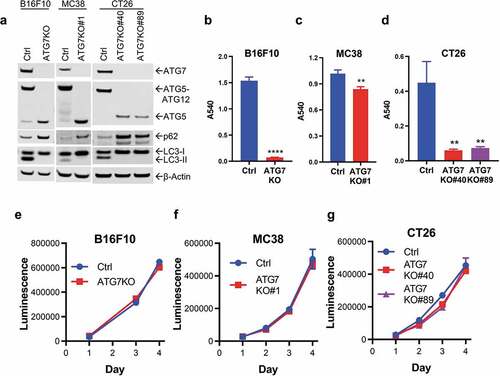
Figure 2. Murine tumors have differential reliance upon ATG7 when grown in immuno-competent hosts.
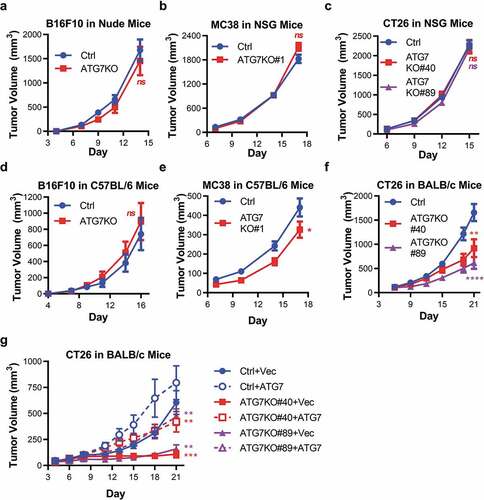
Figure 3. CD8+ and CD4+ T cell contribution to cancer-cell dependence on ATG7 in vivo.
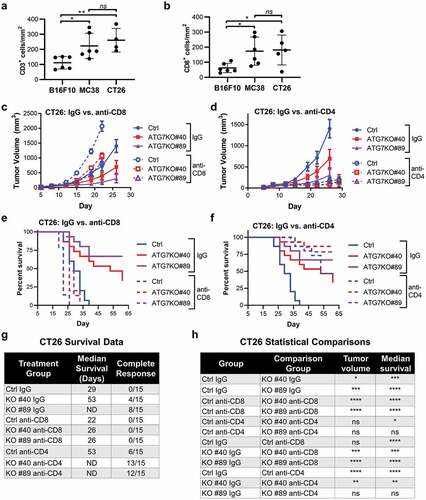
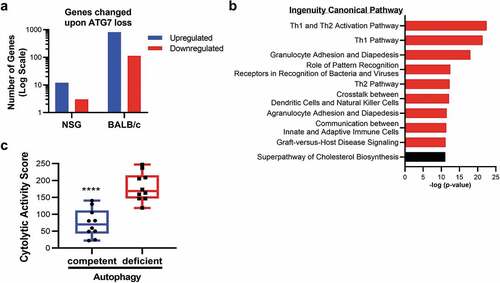
Figure 4. Gene expression and pathway alterations upon Atg7 loss.