Figures & data
Figure 1. Modified vaccinia Ankara (MVA) vaccine-mediated immunogen expressions and the immunogenicity. (a) Schematic of modified vaccinia Ankara (MVA) vaccine used in this study. The MVA vaccine is armed with a fusion gene of sPD1-MUC1-S8. The sPD1 protein is used to enhance the cross-presentation of antigens; MUC1 contained 33 repeats of VNTR; S8 is a deleted form of survivin without the first seven amino acids. (b-c) Confocal images and flow cytometry analysis of MUC1 and survivin expression after intracellular immunostaining and BHK tk-ts13 cells at 24 h post-MVA infection. Scale bar represents 50 µm. (d) Western blotting analysis of MUC1 and survival rates in BHK tk-ts13 cells at 24 h post-MVA infection. (e) Immunization regime illustration; total of four to five mice were in each group. (f, g) Representative images of ELISpot and quantification of ELISpot SFUs in groups. (h) Cytotoxicity evaluation of MVA vaccine-immunized mice (E: T, effector: target cells ratio). N.s., nonspecific protein; M: MUC1 protein; S, survivin protein. Groups drawn in colors: Vehicle (PBS; black), 2MVA-1 (red), 2MVA-2 (brown), 2MVA-3 (pink), 3MVA (blue). One-way ANOVA followed by LSD analysis was performed to analyze the significant differences between groups. *P < .05; **P < .01; ***P < .001
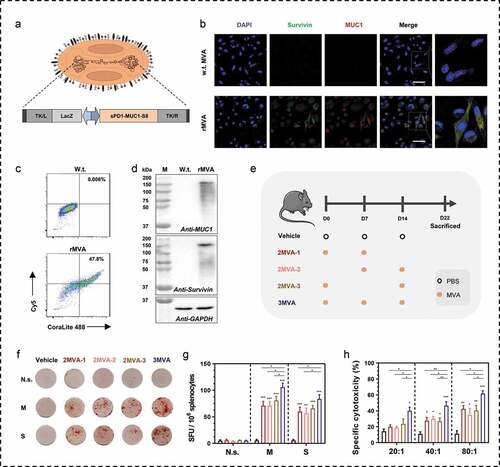
Figure 2. Therapeutic efficacy of modified vaccinia Ankara (MVA) vaccine. (a) Schematic of the therapeutic regime. A total of 1 × 105 MSf+ Lewis cells were injected subcutaneously on the right back of C57/BL/6 mice on day 0, and vaccinations began 4 days after tumor inoculation. (b) Tumor growth curve in groups; each group involved nine to ten mice. (c) Tumor growth curve of each mice in groups. (d) Survival curve; each group involved ten mice. Groups drawn in colors: Vehicle (PBS; black), 2MVA-1 (red), 2MVA-2 (brown), 2MVA-3 (pink), 3MVA (blue). One-way ANOVA followed by LSD analysis was performed to analyze the significant differences between groups. *P < .05; **P < .01; ***P < .001
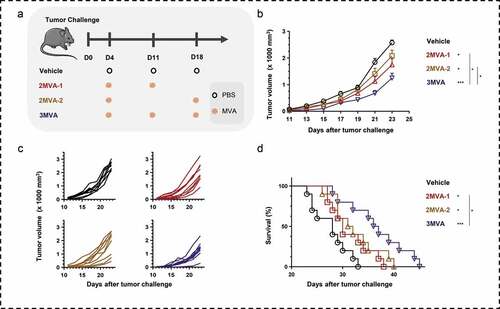
Figure 3. Binding antibodies and neutralizing antibodies against MVA vectors after immunizations. (a) Schematic of rMVA vaccination regime and time points of blood collection. (b) Serum antibody against MVA vector was detected by ELISA. The serum was diluted from 1:100 to 1:12500. (c) Cell viability of BHK tk-ts13 cells at 48 h post-infection of serum-incubated rMVA. The serum was used at the dilution of 1:40 to 1:2560. (d) Representative morphology images of BHK21 tk-ts13 cells at 48 h post-infection of serum-incubated rMVA. Groups drawn in colors: serum before immunizations (Day −1; black), serum after the first immunization (Day 6; blue), serum after the second immunization (Day 13; red), serum after the third immunization (Day 21; brown). One-way ANOVA followed by LSD analysis was performed to analyze the significant differences between groups. Scale bar represents 50 μm. *P < .05; **P < .01; ***P < .001
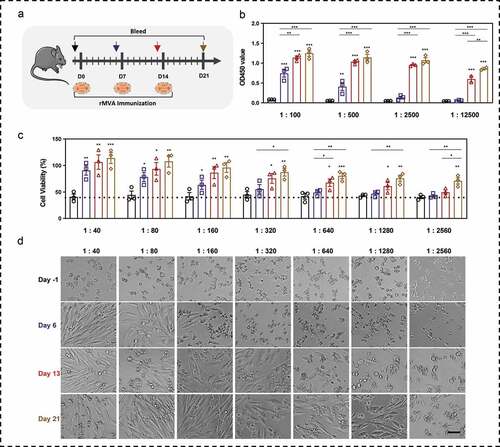
Figure 4. Immunogenicity evaluation of DNA prime-modified vaccinia Ankara (MVA) boost strategy. (a) Schematic of the vaccination regime; total four to five mice in each group. (b, c) Representative images of ELISpot and quantification of ELISpot SFUs in groups. (d, e) Percentage of proliferative CD4+ T (CD4+Ki67+) or proliferative CD8+ T (CD8+Ki67+) in splenocytes after stimulation with survivin or MUC1 protein. (f) Cytotoxicity evaluation of MVA vaccine-immunized mice (E: T, effector: target cells ratio). N.s., nonspecific protein; M: MUC1 protein; S, survivin protein. Groups drawn in colors: Vehicle (PBS; black), 3MVA (red), DNA-1 (brown), DNA-2 (pink), D/M (blue). One-way ANOVA followed by LSD analysis was performed to analyze the significant differences between groups. *P < .05; **P < .01; ***P < .001
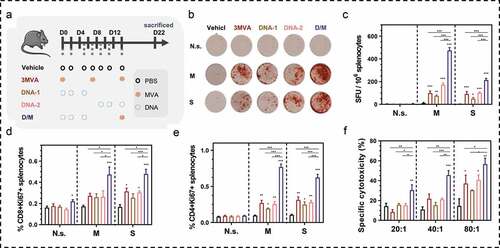
Figure 5. Comparison of therapeutic efficacy among the different heterologous prime-boost strategies. (a) Schematic of the therapeutic regime. 1 × 105 MSf+ Lewis cells were injected subcutaneously on the right back of C57 bl/6 mice on day 0, and vaccinations began 4 days after tumor inoculation. (b) Tumor growth curve in groups; each group included ten mice. (c) Tumor growth curve of each mice in groups. (d) Survival curve; each group involved nine mice. (e–g) Percentage of intra-tumoral proliferative CD4+ T (CD4+Ki67+), proliferative CD8+ T (CD8+Ki67+) and Treg (CD4+CD25+Foxp3+) were analyzed using flow cytometry. (h) The ratio of CD8+Ki67+ T cells to Tregs. (i) Analysis of intra-tumoral MDSC. (j, k) The percentage of MDSC (CD1b+Gr-1+), TAM-M1 (F4/80+CD206−), and TAM-M2 (F4/80+CD206+) was analyzed using flow cytometry. (l, m). Relative mRNA expression of Gzmb and Th1/Th2 cytokines was analyzed using qRT-PCR. Groups drawn in colors: Vehicle (PBS; black), 3MVA (red), DNA-1 (brown), DNA-2 (pink), D/M (blue). One-way ANOVA followed by LSD analysis was performed to analyze the significant differences between groups. *P < .05; **P < .01; ***P < .001
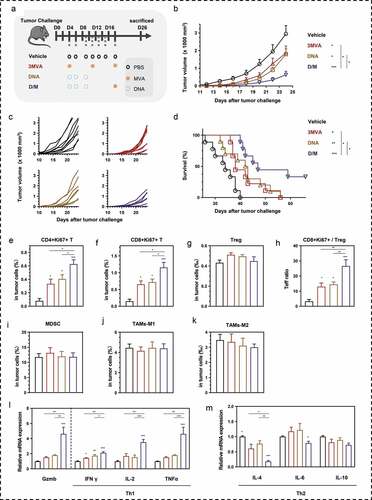
Figure 6. Antigens specific immune memory analysis in (CR) mice. (a) Three CR mice from D/M immunized group were inoculated with B16 cells (3 × 105 cells) on the left back and MSf+ B16 cells (3 × 105 cells) on the right back, after they survived from secondary MSf+ Lewis tumor attack. (b) Growth curve of tumors on both sides of naïve mice or CR mice. (c) The representative images of tumor formation in CR mice or naïve mice at day 25 post tumor inoculations on both sides. (d) Representative images of ELISpot and quantification of ELISpot SFUs in groups. (e, f) Percentage of proliferative CD4+ T (CD4+Ki67+) or proliferative CD8+ T (CD8+Ki67+) in splenocytes after stimulation with survivin or MUC1 protein. Groups drawn in colors in (a-f): Naïve mice (black), CR mice (red). (g, h) representative results and percentage of TCM and TEM in each CR mice analyzed by flow cytometry; Mock, no stimulation; N.s., nonspecific protein stimulation; M, MUC1 protein stimulation; S, survivin protein stimulation. One-way ANOVA followed by LSD analysis was performed to analyze the significant differences between groups. *P < .05; **P < .01; ***P < .001
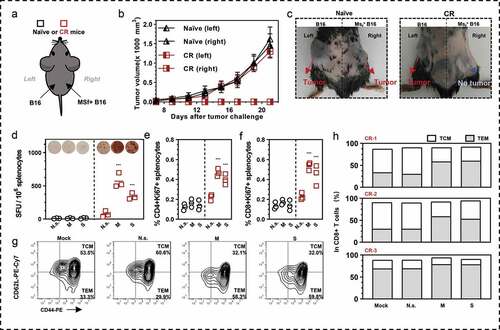
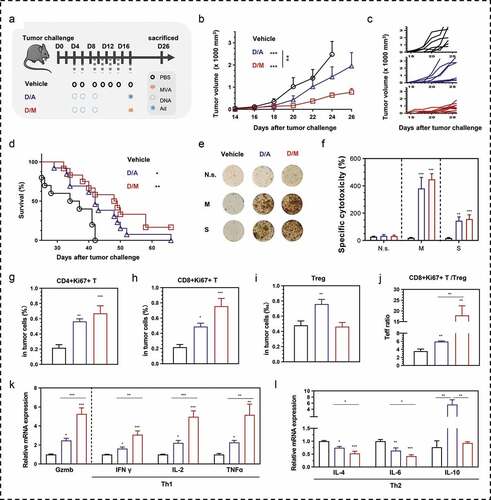
Figure 7. Comparison of therapeutic efficacy between DNA- modified vaccinia Ankara (D/M) and DNA-adenovirus (D/A) prime-boost strategies. (a) Schematic of the therapeutic regime. 1 × 105 MSf+ Lewis cells were injected subcutaneously on the right back of C57 bl/6 mice on day 0, and vaccinations began 4 days after tumor inoculation. (b) Tumor growth curve in groups; each group involved ten mice. (c) Tumor growth curve of each mice in groups. (d) Survival curve; each group included nine mice. (e, f) Representative images of ELISpot and quantification of ELISpot SFUs in groups. (g–i) Percentage of intra-tumoral proliferative CD4+ T (CD4+Ki67+), proliferative CD8+ T (CD8+Ki67+) and Treg (CD4+CD25+Foxp3+) were analyzed using flow cytometry. (j) The ratio of CD8+Ki67+ T cells to Tregs. (k, l) Relative mRNA expression of Gzmb and Th1/Th2 cytokines were analyzed using qRT-PCR. N.s., nonspecific protein stimulation; M, MUC1 protein stimulation; S, survivin protein stimulation. Groups drawn in colors: Vehicle (PBS; black), D/A (blue), D/M (red). One-way ANOVA followed by LSD analysis was performed to analyze the significant differences between groups. *P < .05; **P < .01; ***P < .001