Figures & data
Figure 1. Proliferation of mucosal epithelial cells is increased and p27kip1 expression is decreased in the colon and T cells of Smad4TKO
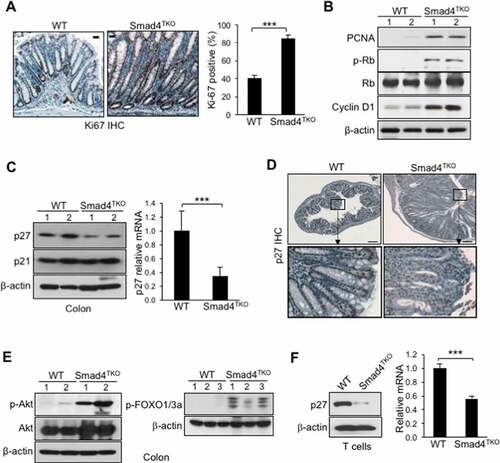
Figure 2. Deletion of p27kip1 in Smad4TKO mice accelerates colitis-associated colon cancer. A) Survival curves of WT, Smad4TKO, p27KO and Smad4TKO/p27Kip1-/- (DKO) mice. B) Photographs of colon, stomach and duodenum from each genotype at 3 months of age. C) Colon weight per length (g/cm) (n = 9). D) Hematoxylin and eosin (H&E) staining of the colon of each genotype at 3 months of age. Scale bar = 100 µm. E) Photograph and mucosal histology of the colon from DKO mice at 3 months of age. Paraffin-embedded sections were stained with H&E. Scale bar = 1 cm. F) Percentage of tumor-bearing mice (n = 10). G) Tumor size (n = 7). H) Tumor numbers per mouse at 3 months of age were determined using a digital eyepiece and an imageJ (n = 7). Error bars indicate S.E.; ***P < .001, **P < .01 compared with each genotype such as WT, p27KO and Smad4TKO
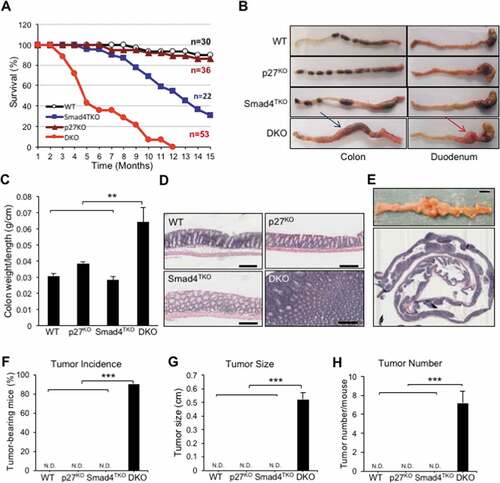
Figure 3. Proliferation of mucosal epithelial cells is increased in the colon of DKO mice. A) Immunohistochemistry (IHC) analysis for Ki-67 in the colon of WT, p27KO, Smad4TKO and DKO mice. The percentage of Ki-67 positive cells among all colon epithelial cells was determined in the colon at 3 months of age using a digital eyepiece. Data represent average ± S.E. (n = 4). B) Expression of PCNA and, p-Rb, and β-actin was measured by Western blot. Results shown are representative of 3 separate experiments. ***P < .001. Scale bar = 100 µm
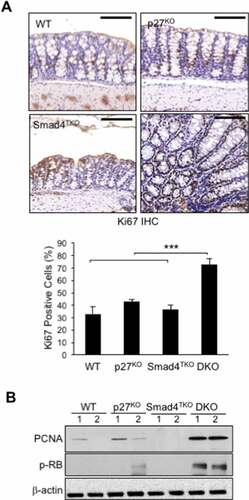
Figure 4. The population of effector memory CD4+ cells in each genotype. A) Analysis of total spleen cells and CD4+ T cells of each genotype at 4 months of age. Cell suspensions were prepared from spleens by filtering through nylon mesh. The cell numbers were counted by trypan blue exclusion assay. The population and number of CD4 T cells in each genotype. Splenocytes were stained with antibodies for CD4 and CD8. Events are gated on the live lymphocyte gate, based on forward and side light scatter (FSC X SSC) by FACS analysis. Bar graphs show the number of spleen cells and the proportion and absolute number of CD4 T cells, respectively. B) Analysis of memory markers (CD44High and CD62LLow) in CD4+ T cell compartment of each genotype. The bar graphs show the proportion and absolute number of CD4 TEM cells from the same experiment depicted in Supplementary Figure 2B. C) Analysis of effector CD4+ T cells expressing pro-inflammatory cytokines of each genotype. Splenocytes per genotype were collected, stained on cell surface with antibody for CD4 and intracellularly with IFN-γ or TNF-γ antibody, and analyzed on CD4+ T cells by FACS. The bar graphs are from the same experiment depicted in Supplementary Figure 2 C. D) IHC staining for CD3+ T cells in colon of each genotype. E) Colon lamina propria per each genotype at 4 months of age were collected, stained with antibodies for CD4 and CD8, and analyzed for effector memory markers (CD44High and CD62LLow) in CD4+ T cell compartment. Bar graphs show the proportion and absolute number of CD4 TEM cells and effector CD4 T cells producing IFN-γ, respectively. ***P < .001, **P < .01, *P < .05. Scale bar = 200 µm
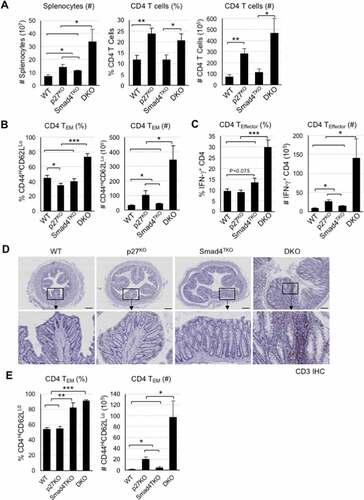
Figure 5. Decreased percentage of natural Foxp3+ regulatory T cell (nTreg) and impaired TGF-β-induced Foxp3+ Treg (iTreg) in DKO mice. A) Analysis of nTreg population in CD4+ T cell compartment of each genotype mice. Splenocytes per genotype at 4 months of age were collected, stained on cell surface with antibody for CD4 and intracellularly with Foxp3 antibody, and analyzed on CD4+ T cells by FACS. Events are gated on the live lymphocyte gate, based on forward and side light scatter (FSC X SSC). Bar graph represents percent nTreg from each genotype at 4 months. B) Analysis of iTreg population in CD4+ T cell compartment of each genotype. Cells from Spleen per genotype at 2 months of age were collected and incubated with anti-CD3/CD28 for 72 hrs with (gray bar) or without (black bar) TGF-β (2 ng/ml) and intracellularly stained with Foxp3 antibody. Results are representative of three independent experiments using pooled spleens from two mice per genotype. Bar graph of percent iTreg from each group. C) Analysis of suppressive capacity of iTreg from CD4+ T cell compartment of WT and Smad4TKO. CD4+CD25+ iTregs from panel B were co-incubated with naïve CD4 T cells with 1 μg/ml of Con A for 3 days and the proliferation of naïve CD4 T cells was measured by H3 thymidine incorporation for the final 16 hrs of culture. ***P < .001, **P < .01, *P < .05
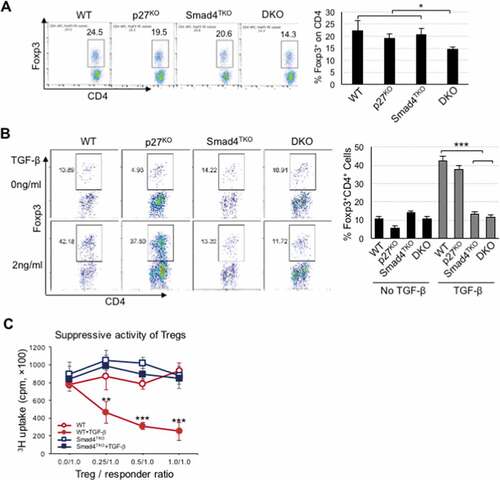
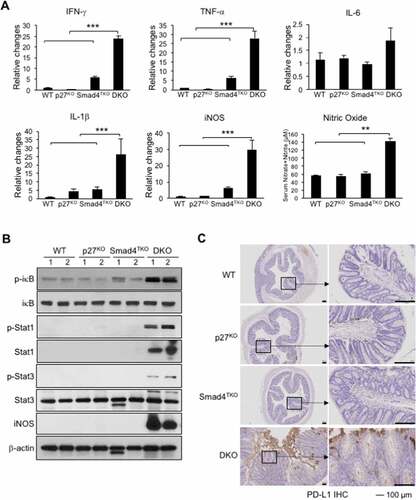
Figure 6. Increased inflammatory responses in the colon of DKO mice. A) IFN-γ, TNF-α, IL-6, IL-1β and iNOS were measured by real-time PCR in colon mucosa of WT, p27KO, Smad4TKO and DKO mice at 3 month of age. Nitric oxide (nitrate + nitrite) concentration in sera of each genotype at 3 months of age. B) Expression of phospho-iκb, phospho-Stat1, phospho-Stat3, and iNOS was determined in colon epithelia of each genotype (3 month old) by Western blot analysis. β-actin was used as the loading control. C) Immunohistochemistry (IHC) staining for PD-L1 in colons of each genotype. Scale bar = 100 µm