Figures & data
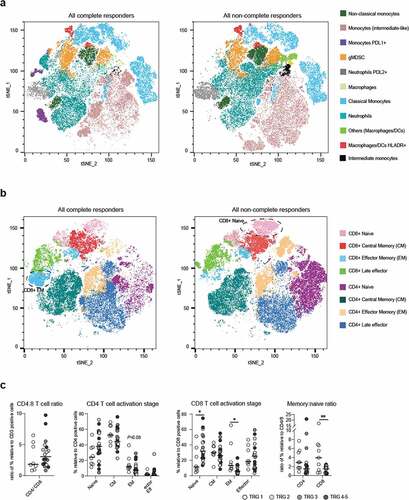
Figure 1. Multicolor IHC shows increased T cell infiltrate and importance of spatial distribution in the tumor of complete responders
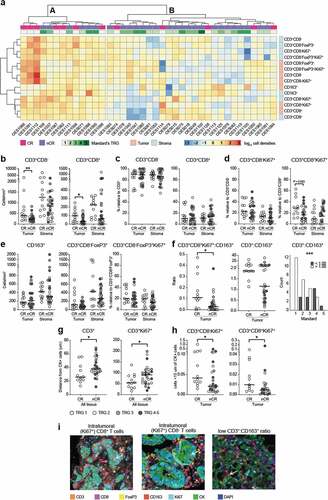
Figure 2. Flow cytometry on pre-treatment biopsies did not identify any response-specific immune signatures
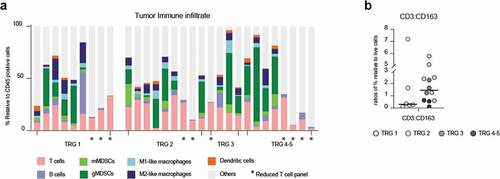
Figure 3. NanoString analysis shows an more cytotoxic cells complete responders than non-responders
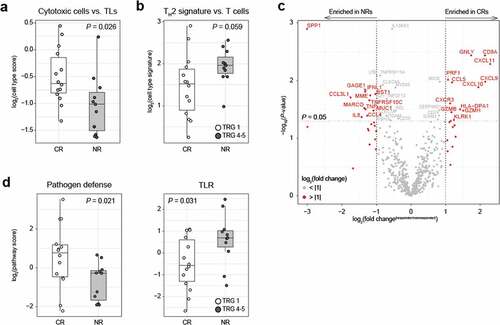
Figure 4. Flow analysis on circulating cells shows enriched CD8+ memory T cells in complete responders