Figures & data
Table 1. Clinical characteristics of individuals recruited into this study. Tumor grade refers to the WHO glioma grade, and IDH refers to the isocitrate dehydrogenase mutation, both of which were determined by a pathologist. * – data from this individual were not included in further analysis
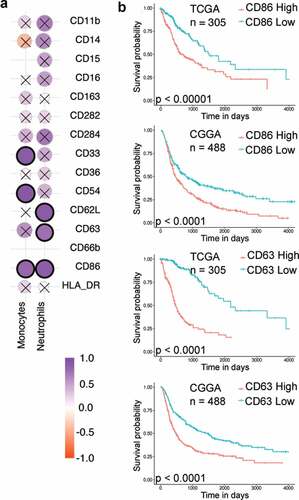
Figure 1. Immune cell frequencies
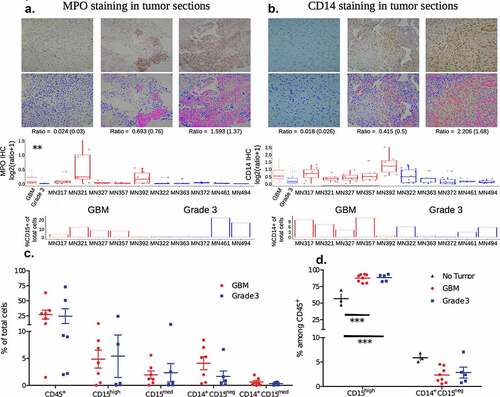
Figure 2. Clustering analysis of flow cytometry data
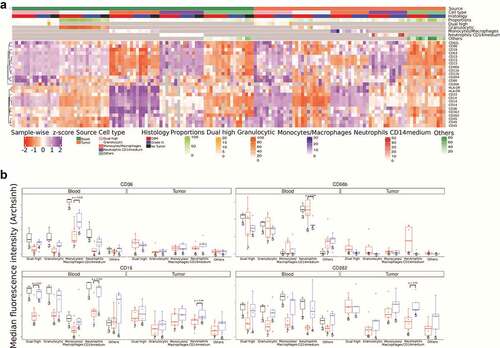
Figure 3. Neutrophil proteomics
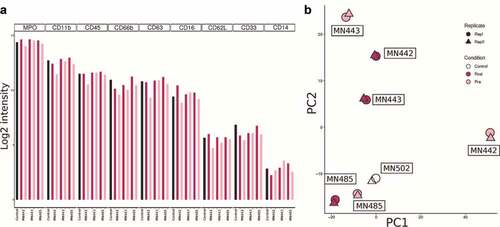
Figure 4. Expression of CD163
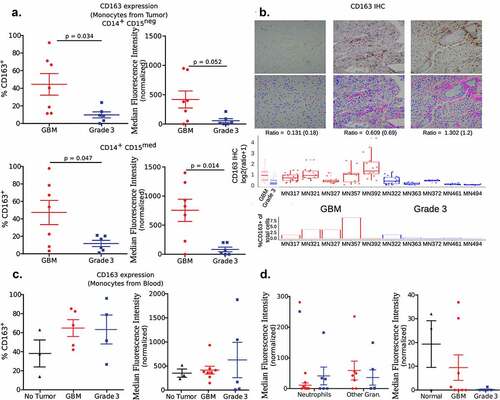
Figure 5. Correlation analysis