Figures & data
Figure 1. Overview of the IHC and PBMC cohorts. In this study, tumor samples of 81 prostate cancer patients were used for IHC. If available, multiple tumor samples per patients were analyzed, including tissue of the hormone-sensitive and castrate-resistant setting. For TCR sequencing, PBMCs of 48 patients were used. Patients were classified into four genomic subgroups as depicted in the figure. There were 15 HRD patients in the IHC cohort and 16 in the PBMC cohort.
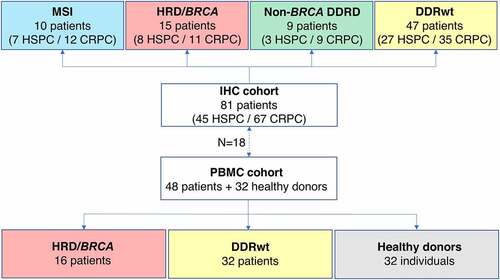
Figure 2. Tumor-infiltrating lymphocyte density per tissue site. A. Multiplex immunohistochemistry (IHC) images of the four most prevalent tissue sites (≥ 5 samples included). The scalebar represents a length of 100 µm. Additional representative IHC images are shown in supplementary figure 3. B. CD3+, CD8+, CD3+CD8−FoxP3− and FoxP3+ cell density in the tumor (upper panel) and stroma compartment (lower panel) per tissue site. For all cell subsets, we observed significant differences between tissue sites, both in the stromal and tumoral compartments (Kruskal-Wallis test, p ≤ .05). Black lines indicate significant differences between pairs (Dunn’s test, * p ≤ .05; ** p ≤ .01; *** p ≤ .001). In some patients, no T cells were present. To enable visualization of cell densities on a log scale, the T cell densities of these patients were replaced by 0.5 cells/mm2 (~lowest value in the plots). Differences between paired samples are displayed in supplementary figure 4.
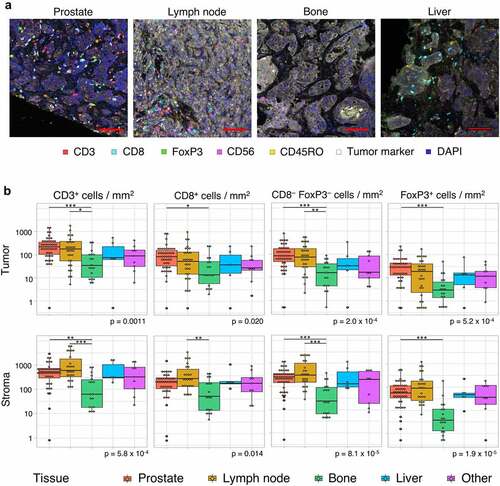
Figure 3. Intratumoral TIL density per tissue site. Intratumoral TIL densities in prostate tissue (a), lymph node metastases (b) and other tissue sites (c) are displayed. The stacked bar plot at the left shows the absolute T cell densities per sample, subdivided into CD8+, CD3+CD8−FoxP3− and FoxP3+ cells. Samples are ordered based on total CD3+ T cell density. The bars below provide information on genomics subgroup, androgen sensitivity and tissue site. At the right, T cell densities per genomic subgroup as displayed in boxplots. In some patients, no T cells were present. To enable visualization of cell densities on a log scale, the T cell densities of these patients was replaced by 0.5 cells/mm2 (~lowest value in the plots).
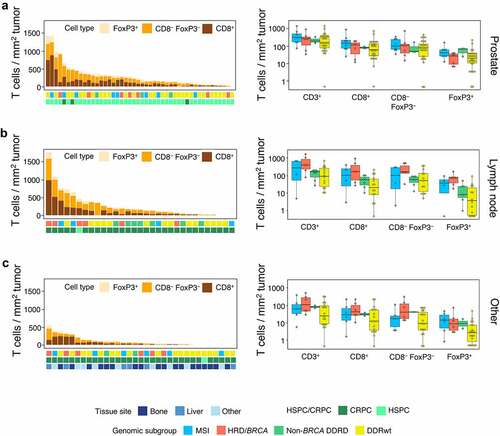
Figure 4. PD-L1 expression. (a) Examples of PD-L1 stainings. The left image shows a tumor sample with PD-L1+ immune cells in 1% of tumor area and PD-L1 expression on 1% of tumor cells. The sample with the highest tumor cell expression in our cohort is depicted on the right. In this sample (only a part is shown) 30% of tumor cells expressed PD-L1 on their cell surface. The scalebar represents a length of 100 µm. (b) The proportion of patients with PD-L1 expression on immune (left) and tumor cells (right) per genomic subgroup. To avoid overrepresentation of patients with multiple samples, samples were weighted based on the number of available samples per patient (with each sample counting as 1 observation if one sample was available and each sample counting as 0.5 observation if two samples were available). No significant differences were observed between genomic subgroups.
Figure 5. Peripheral T cell receptor (TCR) repertoire. (a) TCR diversity and clonality per genomic subgroup. (b) TCR diversity and clonality in patients who have received only 0–1 lines of prior systemic therapy. This analysis includes 6 patients with HRD/BRCA and 27 patients with DDRwt. (c) T cell cluster analyses. As the HRD/BRCA subgroup contained two times less individuals than the DDRwt and HD group, we split the DDRwt and HD cohorts in two and performed two independent experiments, comparing the same cohort of HRD/BRCA patients with different cohorts of DDRwt patients and HDs. We focused on the clusters that were shared between HRD/BRCA and DDRwt patients or were exclusive to these groups (n = 2232 in experiment 1; n = 2164 in experiment 2). Fold-changes were calculated by dividing the median frequency of a cluster in the HRD/BRCA subgroup by the median frequency of the same cluster in the DDRwt subgroup. Clusters that were exclusively present in either the HRD/BRCA (far right, orange dots) or DDRwt (far left, blue dots) were given artificial log2 fold-change values for plotting purposes. In total, 84 clusters were exclusive to HRD in both experiments compared to 33 in the DDRwt subgroup (supplementary file).
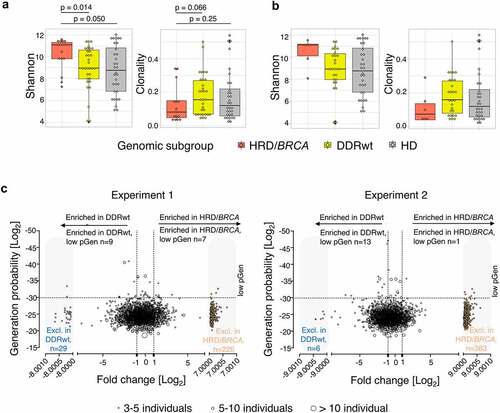
Table 1. Response to ICIs by HR status.
