Figures & data
Figure 1. Differences in vaccine effectiveness by irradiated whole tumor cells.
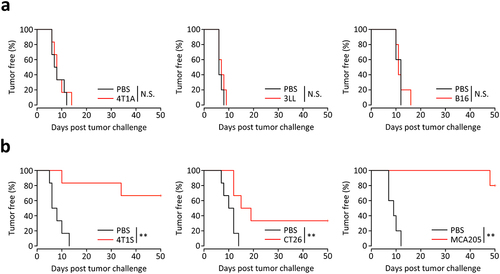
Table 1. Highly expressed genes in vaccine-effective cancer cells after radiation.
Table 2. Interferon family gene expression in each radiated cancer cell.
Figure 2. Vaccine effectiveness by each irradiated transgenic whole tumor cell.
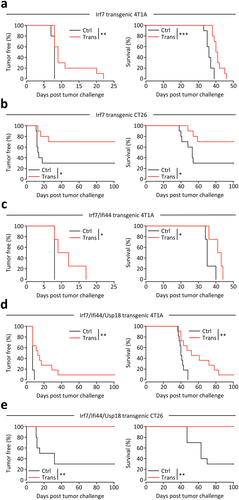
Figure 3. Effects of Irf7 on vaccine effectiveness by radiated whole tumor cells. .
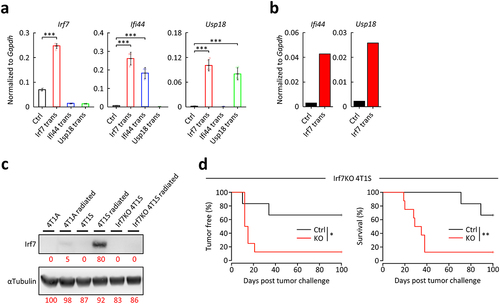
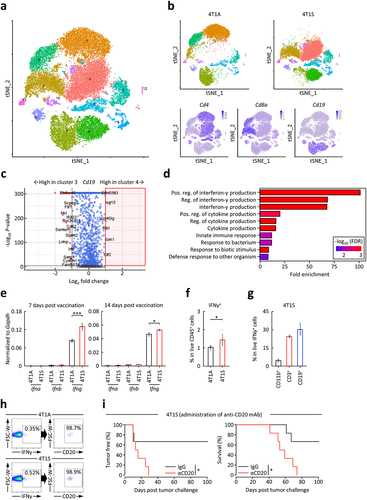
Figure 4. Effects of IFNγ+ B cells on vaccine effectiveness by irradiated whole tumor cells.
Supplemental Material
Download Zip (8.2 MB)Data availability statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
