Figures & data
Figure 1. Scheme illustrating the ability of virus particles to reach active binding sites in columns packed with porous particles (A) and CIM monolithic columns (B).
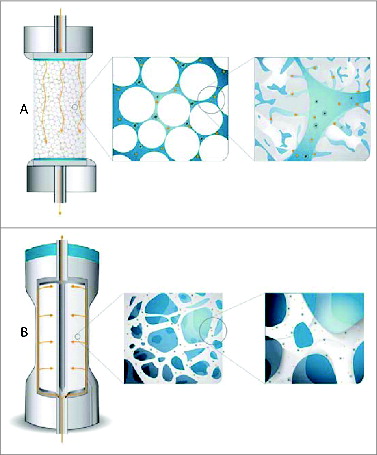
Figure 2. Flow independent resolution of CIM monolithic columns; gradient elution of the mixture of 6 proteins on CIMac SO3 Analytical ColumnTM at the flow rates 0.5, 1.0 and 2.0 mL/min. Column: CIMac SO3 Analytical ColumnTM (5.2 mm I.D. × 5.0 mm); Sample: Test protein mixture (1) myoglobin, (2) trypsinogen, (3) ribonuclease A, (4) α-chymotrypsinogen A, (5) cytochrome C, (6) lysozyme; Injection volume: 10 μl; mobile phase A: Buffer A: 20 mM Na-phosphate, pH 6.0, Mobile phase B: Buffer B: 20 mM Na-phosphate + 1.0 M NaCl, pH 6.0, Gradient: a linear gradient from 0 to 28% buffer B in 30 CV, Detection: UV at 280 nm, HPLC system: Knauer high pressure gradient HPLC system.
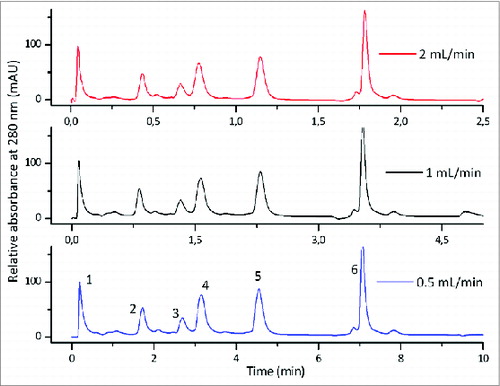
Table 1. Dynamic binding capacity and virus recovery on different chromatography supports for different virus (-like) materials
Figure 3. General scheme of virus production process with emphasis on DSP part. Abbreviations: UF-ultrafiltration; DF-diafiltration; IEX-ion exchange; HIC-hydrophobic interaction mode; SEC-size exclusion chromatography.
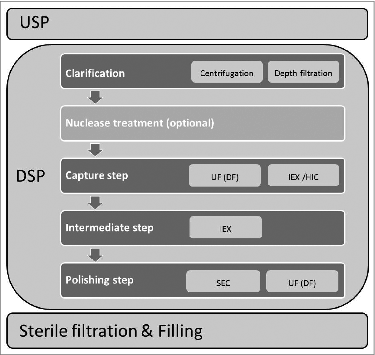
Figure 4. Analysis of crude T4 lysate collected during fermentation: (A) between 0 and 95 min; (B) between 120 and 205 min; (C) between 215 and 325 min. Conditions: Stationary phase: 0.34 mL CIM DEAE disk monolithic column; Mobile phase: Buffer A: 20 mM Tris, pH 7.5. Buffer B: 20 mM Tris, 1.0 M NaCl, pH 7.5; Gradient: linear gradient from 0 to 100% buffer B in 1.5 min; Flow rate: 4 mL/min; Injection volume: 1 mL; Detection: UV at 280 nm; reused from Smrekar et. al.Citation44.
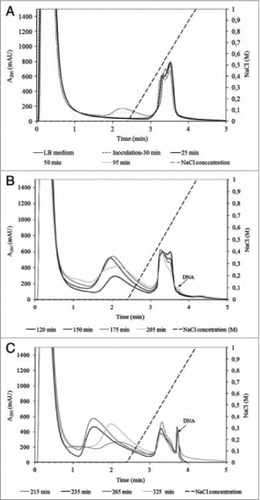
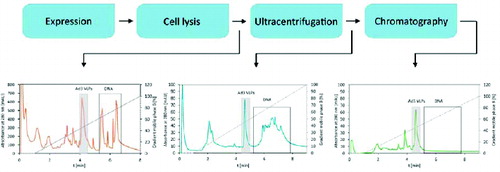
Figure 5. Fingerprint HPLC elution profiles of Ad3 VLPs purification process. Elution profiles depict the purity/impurity profiles of samples containing Ad3 VLPs after particular DSP steps.