Figures & data
Table 1. Complete Ab genomes from NCBI used in this study
Figure 1. Phylogenetic relationship of Acinetobacter baumannii. The phylogenetic lineage of fully sequenced Ab strains analyzed in this study was inferred by the neighbor-joining method performed in MEGA6 using the strategy previously published in.Citation35
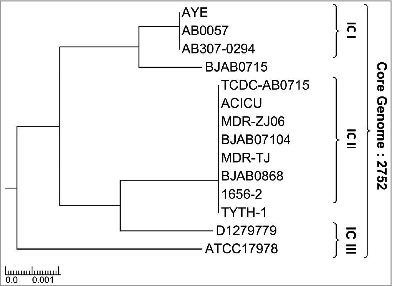
Figure 2. Schematic summarizing the flow chart of Reverse Vaccinology used in this study. 1.Comparison of genomic sequences to obtain the orthologous core genes by the in silico web-based tools (MBGD). 2. 2.1 PSORTb 3.0 to predict the cellular localization of each protein. 2.2 Outer membrane and extracellular proteins identification by proteomics analyses/references. 3. Blasting of protein sequences and literature-review to determine if the proteins have ever been used as immunogens. 4. Expression and purification of the consensus sequence of candidate immunogens. 5. Utilization of the mouse pneumonia model to assess vaccine immunogenicity and protective ability against A. baumannii infection. 6. Development of a broadly protective A. baumannii vaccine formulation.
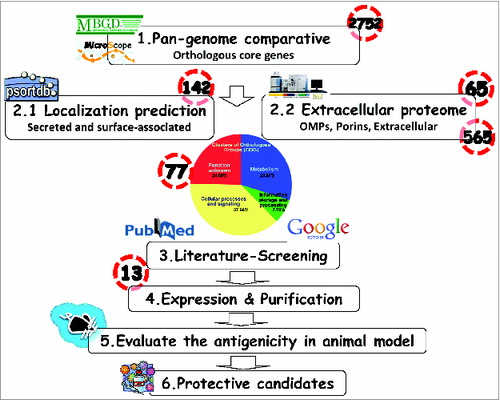
Table 2. Most promising immunogens for an A. baumannii vaccine
Figure 3. 12% SDS-PAGE and Western blot of purified recombinant proteins. The purification process was performed using immobilized-metal affinity chromatography. (A) Lane M-standard protein markers, Lane 1-Whole bacterial lysate after induction of OmpK (27.6 kDa) by IPTG, Lane 2-purified recombinant OmpK protein, Lane 3-Whole bacterial lysate after induction of Ompp1 (50.6 kDa) by IPTG, Lane 4-purified recombinant Ompp1 protein, Lane 5-Whole bacterial lysate after induction of FKIB (25.6 kDa) by IPTG, Lane 6-purified recombinant FKIB protein. (B) Western blot analysis of purified recombinant protein using mouse anti-His antibody.
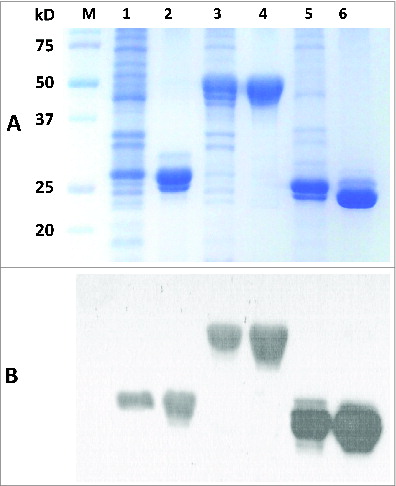
Figure 4. Mouse immunogenicity studies of individual antigens. (A) Groups of C57BL/6 mice (n = 4) were intramuscularly immunized with either 3 or 10 μg of individual antigen (OmpK, FK1B and Ompp1) formulated with CFA/IFA adjuvant, or PBS on day 0, 14 and 21. At day 42, blood samples were collected and tested against each immunogen. Serum IgG antibody titers are reported. (B) Survival curves of C57BL/6 mice immunize with Ommp1. Groups of C57BL/6 mice (n = 5) were subcutaneously immunized with either 3 or 30 μg of Ommp1 formulated with CFA/IFA adjuvant, or PBS on day 0, 14 and 21. At day 42, the mice were challenged with 5 × 107 CFU of freshly grown A. baumannii ATCC17978. The survival rates of mice were recorded.
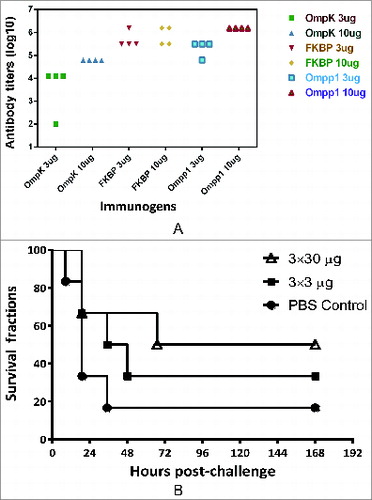
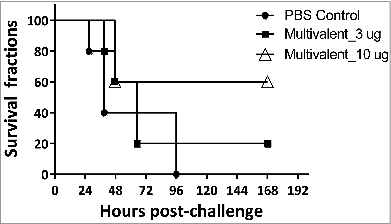
Figure 5. Survival curves of C57BL/6 mice immunized with multivalent vaccines Groups of C57BL/6 mice (n = 5) were subcutaneously immunized with multivalent antigens or PBS on day 0, 14 and 21. At day 42, the mice were challenged with 1.34 × 108 CFU of freshly grown A. baumannii ATCC17978. The survival rates of mice were recorded.