Figures & data
Figure 1. Endothelial cells (ECs) in cultures. (A) Representative human microvascular ECs (HMECs) isolated from adipose tissue and used to prepare the SANTAVAC preparation. HMECs had numerous cytoplasmic extensions and/or a cobblestone-like morphology typical of adipose-derived microvascular ECs.Citation89 Images were obtained using a Leica DM5000B microscope. Cells were isolated by using magnetic beads linked to anti-CD31 antibodies (visible on the cells). (B) Example of ECs sprouting around cancer cells under angiogenic stimuli. Artwork prepared using data obtained from a multiphoton microscopy image of a tumor spheroid in a collagen matrix.Citation90
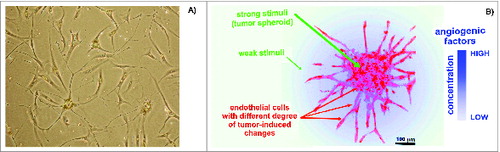
Figure 2. Cell proteomic footprinting. (A) Adherent cell culture after washing away traces of culture medium and subsequently treated with a protease. Released fragments of the cell surface proteins were collected and subjected to mass spectrometry analysis. The set of obtained peptide molecular weights represents the cell culture proteomic footprint. (B) Examples of cell proteomic footprints for non-ECs (MCF-7 and HepG2) and HMECs induced to grow in the presence of stimuli provided from normal tissue (HMECECGS) or cancer cells (HMECHepG2). Adapted from.Citation37,41
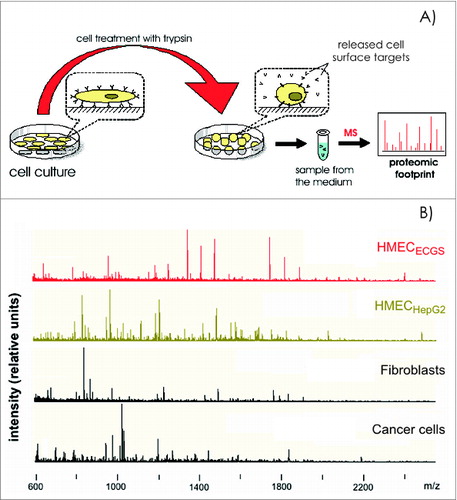
Figure 3. Degree of change in the HMEC surface antigen expression profile after incubation in the presence of tumor-conditioned medium. (A) Principle component analysis (PCA) of cell surface profiles obtained from HMECs and control non-ECs (HepG2 and MCF-7) that were projected in the space of the first 2 principal components. (B) PCA of cell surface profiles obtained only for HMECs projected into the space of the first 3 principal components. Cell surface profiles are shown for HMECs stimulated to grow in the presence of EC growth supplement (1HMECECGS and 2HMECECGS), MCF-7 cell-conditioned medium (1HMECMCF-7 and 2HMECMCF-7), LNCap cell-conditioned medium (1HMECLNCap and 2HMECLNCap), or HepG2 cell-conditioned medium (1HMECHepG2 and 2HMECHepG2). Superscript numbers correspond to different donors used to establish HMEC primary cultures.
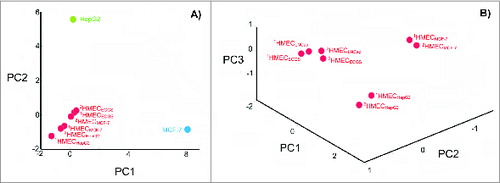
Figure 4. Diagram representing (A) the relative ratio of surface targets that are accessible for humoral and cytotoxic immunity, and the remaining cellular content that is undesired for preparation of cell-based vaccines; and (B) the effects of trypsin impurities and cell death rates on the preparation of cell surface antigens. The lighter region in the lower left corner represents conditions where cell-surface antigen preparations are compatible with the preparation of SANTAVAC vaccines. Trypsin purity is reflected by the levels of enzymatic activity. Adapted from Lokhov.Citation40
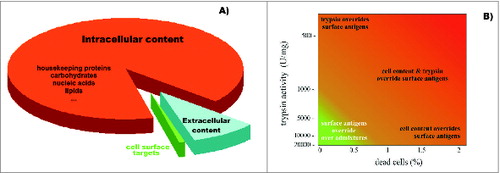
Figure 5. Cytotoxicity assays (CTAs). Data represent the cytotoxicity of effector CTLs against target HMECs, plotted vs. the similarity between surface profiles of target cells and cells used as a source of antigens for eliciting the targeted immune response. Data represent the mean value of 3 independent measurements. The similarity between the cell surface profiles is presented as the correlation coefficient r between corresponding proteomic footprints. 1▸2 – First letter corresponds to HMECs used as a source of antigens for eliciting immune response in the CTA, and the second letter corresponds to the target HMECs used in the same CTA. Red and brown letters correspond to autologous and allogeneic antigens, respectively. Different letters are used to identify HMECs stimulated to grow in the presence of an EC growth supplement (G), MCF-7 cell-conditioned medium (M), LNCap cell-conditioned medium (L), or HepG2 cell-conditioned medium (H). Data were scaled to bring all controls to equal values (25,000 cells, see CONTROL line). Dashed lines show examples of linear dependence (n = k*r + b) between CTA data n and r. All data in the plot were described by linear equations, and the variations of the coefficients (k and b) were interdependent.Citation37
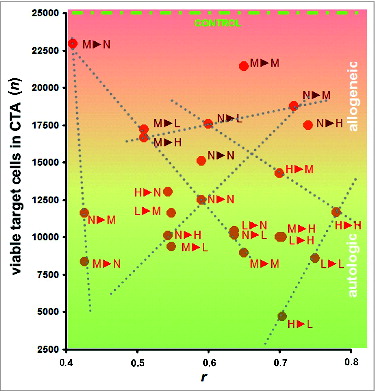
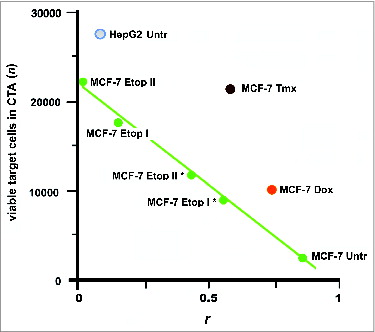
Figure 6. Escape of cancer cells from the immune response in CТA as a result of cell surface profile changes induced by the selective pressure of drug treatment. These changes can lead to a loss of vaccine efficacy based on cancer cell antigens. Points are presented for MCF-7 and HepG2 cells that were untreated (MCF-7 Untr and HepG2) or MCF-7 cells treated as follows: IC96 doses of doxorubicin (MCF-7 Dox) or tamoxifen (MCF-7 Tmx); a single dose of IC96 etoposide (MCF-7 Etop I) or IC50 etoposide (MCF-7 Etop I*); and two separate doses of IC96 etoposide (MCF-7 Etop II) or IC50 etoposide (MCF-7 Etop II*). Linear approximations for MCF-7 Untr and MCF-7 Etop I, II, I*, and II* (green line) are shown. The average number of viable cells in 3 wells is presented. Correlation values (coefficient r) were calculated for MCF-7 and HepG2 cell surface profiles used to generate antigens for eliciting immune response and the surface profile of target MCF-7 cells used in same CTA. Adapted from.Citation47