Figures & data
Figure 1. Analysis of worldwide distribution of HLA alleles and HIV-1 prevalence. (A) HIV-1 subtype distribution in China based on HIV DATABASES (www.hiv.lanl.gov). (B) HLA allele distribution.
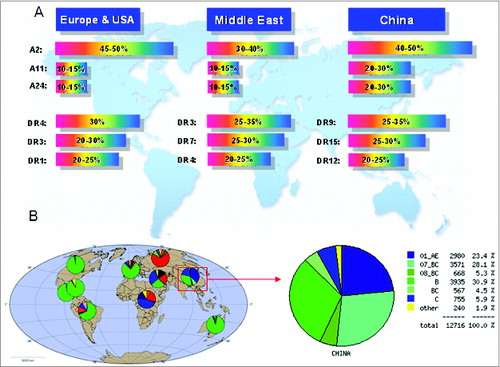
Figure 2. Strategy of HIV-1 multi-epitope DNA vaccine design, including epitope selection, vaccine design, and vaccine evaluation.
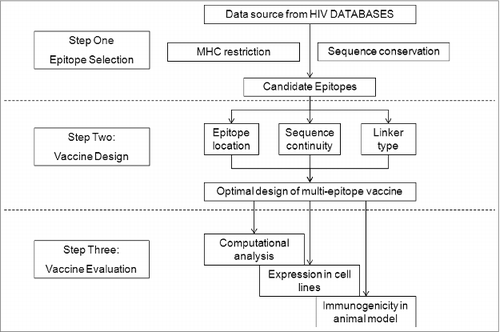
Figure 3. Conservation analysis of the candidate epitopes within HIV-1 viral subtypes prevalent in China. Two hundred and 7eight strains of HIV-1 (52 B subtypes, 160 CRF01_AE subtypes, 33 CRF07_BC subtypes, and 33 CRF08_BC subtypes) prevalent in mainland China were included in this study. The conservative characteristics of AAon the selected epitopes were analyzed by aligning the epitopes with the corresponding molecules on 278 strains of HIV-1, and the conservation was expressed as a percentage of the number of HIV-1 strains having the conserved AAon the epitope from the 278 strains. The core sequences of the CTL and Th epitopes are highly conserved with rare mutations.
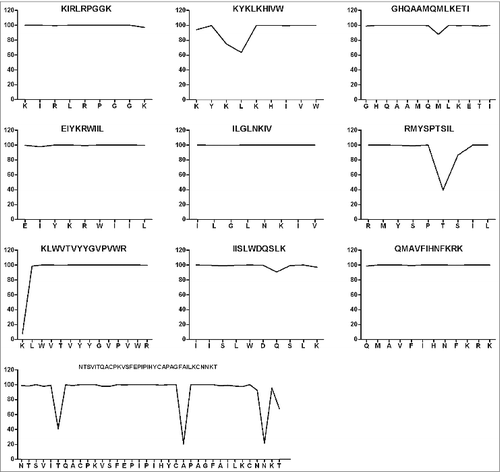
Figure 4. Schematic diagram of the 3 multi-epitope-based DNA vaccines. Epitopes include a 36-aa-long peptide acting as a T-helper epitope (aa 197–232), 6 CTL epitopes in Gag, 2 CTL epitopes in Env, and one epitope in Pol. Epitope locations, continuity, and linkers differ in the 3 sequences. Epitopes are connected using 2 different linkers: GGGS (solid line) was used for pJW4303-MEG1 and pJW4303-MEG2, while AAY (dashed line) was used for pJW4303-MEG3. Numbers below models stand for epitope numbers, which are shown in .
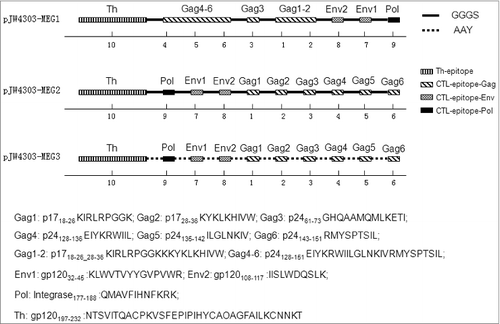
Figure 5. Secondary structure analysis of the multi-epitope-based candidate vaccines. The secondary structure, including α region, β region, turn region and coil region, and antigenicity-related properties, such as hydrophilicity, antigenic index, surface probability, and flexible region, of the artificial proteins based on the conserved epitopes were predicted using PROTEAN pattern of LASERGENE software. Panels A, B, and C are the analytic results of the DNA vaccines pJW4303-MEG1, pJW4303-MEG2, and pJW4303-MEG3, respectively.
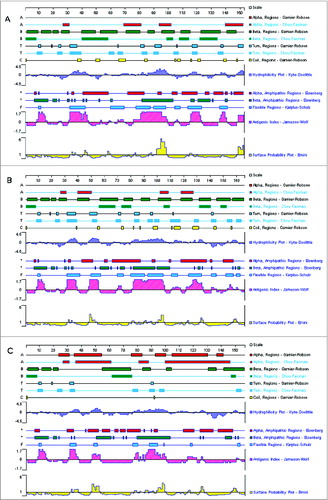
Figure 6. Identification and expression of the DNA vaccines in vitro. (A) Restriction enzyme analysis of the 3 multi-epitope DNA vaccines, with digestion by BamH I and EcoR I,follwed by 1% agarose electrophoresis. Two bands (about 5,000 bp and 500 bp) were observed for all 3 recombinant plasmids. Lane 1, pJW4303-MEG1; lane 2, pJW4303-MEG2; lane 3, pJW4303-MEG3; M, Trans 2K Plus marker. (B) Western blot analysis of DNA vaccine protein expression in vitro. 293T cells were transiently transfected with pJW4303-MEG1, pJW4303-MEG2, or pJW4303-MEG3. Expression of the proteins was identified by Western blot using rabbit anti-MEG1.E polyclonal antibody as primary antibody. A 16 kDa band was observed for both pJW4303-MEG1 and pJW4303-MEG2, but no signal was detected for pJW4303-MEG3 or the control. Lane 1, pJW4303-MEG1; lane 2, pJW4303-MEG2; lane 3, pJW4303-MEG3; lane 4, negative control. Protein marker beside the film used to illustrate protein sizes was cut from the PVDF membrane.
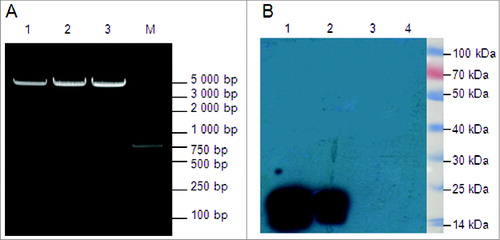
Figure 7. Evaluation of immune responses in BALB/c mice induced by pJW4303-MEG1, pJW4303-MEG2, and pJW4303-MEG3. BALB/c mice were intramuscularly immunized with pJW4303-MEG1, pJW4303-MEG2, and pJW4303-MEG3 at a dose of 10 μg of DNA dissolved in 200 μl of PBS according to the immunization schedule. (A) Two weeks after the final immunization, sera were collected and peptide-specific IgG was measured by ELISA. (B) On day 21 after the final immunization, mice were sacrificed, and splenocytes were stimulated with pool-peptides for 48 hours to detect cellular immune response by ELISPOT assay. (C) Splenocytes from mice injected with pJW4303-MEG1 were also stimulated with H-2d-restricted epitopes to evaluate cellular immune response to single peptides.
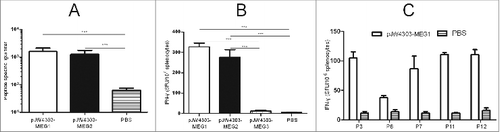
Table 1. Characteristics of the candidate epitopes selected for epitope-based vaccine construction
