Figures & data
Table 1. Representative recombinant VLP-based human vaccines on the market
Figure 1. Presentation of different truncated versions of HEV pORF2. E2s (aa 459–606), the shortest version to form dimer, harbors the major neutralizing epitopes, and its crystal structure was determined with a high resolution of 2.0 Å. p239 (aa 368–606) is the vaccine antigen in Hecolin® in a particulate form.Citation8,9 p495 (aa 112–608), which can form VLP with T = 1 icosahedral symmetry, is used in the other HEV vaccine which has been tested in a phase II clinical trial.Citation17,91 p595 (aa 14–608) also can form a particle (T = 3) which is more similar to the native virions.Citation15 The structure of T = 3 VLP was acquired by using the T = 1 VLP (PDB ID: 2ZTN) as a template to build the homology model with the spatial information of side chain because the reconstruction model from cryo-EM data of T = 3 VLP (PDB ID: 3IYO) only recorded the information of αC atoms. The peptide located at aa 368–459 is supposed to play a key role in inducing multimerization of p239.
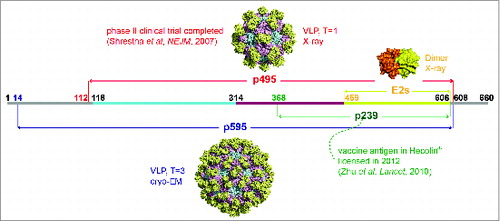
Figure 2. The binding sites of representative neutralizing antibodies on the HEV VLP surface. (A) The pORF2 monomer is divided into 3 sections named the S domain (aa 118–313), the P1 domain (aa 314–453) or P domain (aa 320–455) and the P2 domain (aa 454–606) or P domain (aa 456–606), which are shown in color blue, purple, and yellow, respectively. The P2 or P domain is dimeric and harbors all of the identified neutralizing epitopes. The neutralizing epitopes determined by different methods against several neutralizing antibodies are shown in different colors. Such as E479, D481, T484, Y485, S487, Y532 and S533 for FAB244;Citation17 S487, S488, T489, P491, N562 and T564 for MAB1323;Citation15 D496, G591 and P592 for MAB272;Citation15 E479, Y485, I529 and K534 for 8H3; E479, S497, R512, K534, H577 and R578 for 8C11. These antibodies are useful serological markers for evaluating the clinical efficacy of the vaccine. (B) Key neutralizing epitopes on P (P2) domain of pORF2. The S domain, M (P1) domain and P (P2) domain are colored in blue, purple, and yellow, respectively.
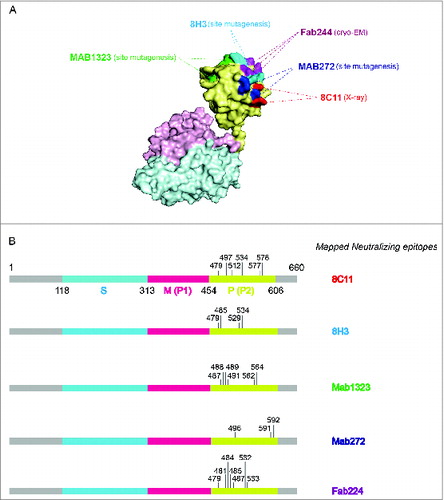
Figure 3. The crystal structure of 8C11Fab in complex with the E2s domain. (A) The structures of the E2s-8C11Fab complexes. (B) The epitope of mAb 8C11 on E2s, including E479, T497, R512, K534, H577 and R578 on the recombinant HEV capsid protein at the interface with an interacting Fab of the 8C11. (C) Interacting residues at the interface between E2s and 8C11Fab. The interactions include hydrogen bonds, Pi-Pi interaction and salt bridge.
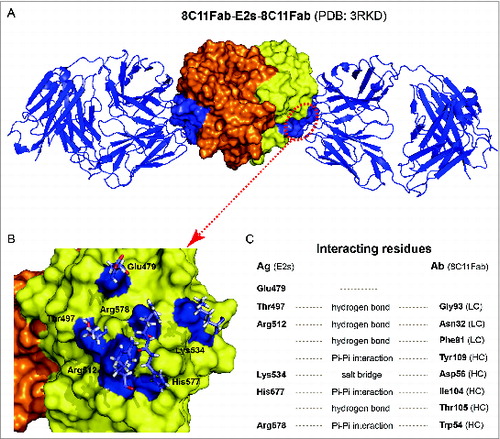
Figure 4. (See previous page) Comparison of different loops, harboring key neutralizing epitopes, of 4 genotypes HPV11, HPV16, HPV18 and HPV35 (for which X-ray structures are available).Citation96 (A) The BC loop, DE loop, EF loop, FG loop and HI loop colored in red, green, blue, yellow and magenta, respectively, are shown in L1 monomer, pentamer and T = 7 VLP. (B) Comparison of the L1 surface loops, BC loop, DE loop, EF loop, FG loop and HI loop respectively. HPV11, HPV16, HPV18 and HPV35 are shown in different colors (blue, magenta, yellow and green, respectively). (C) Localization of the different loops in the full length of HPV16 L1 protein. The epitope regions of 2 type-specific neutralizing mAbs are labeled with an arrow (H16.V5 binding to the FG and HI loop, H11.B2 binding to the DE loop).
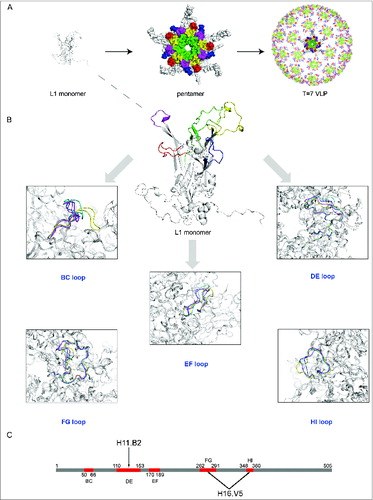
Table 2. Structure determination of recombinant virus-like particles or the subunit antigens of prophylactic vaccines against 3 viruses: HEV, HPV and HBV
Table 3. The key epitope information of neutralizing mAbs against HEV, HPV and HBV
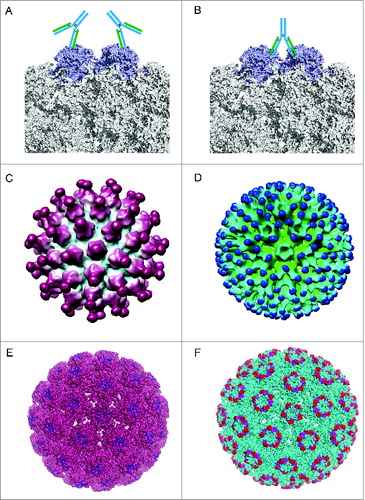
Figure 5. Different binding patterns of antibodies to BPV and HPV.Citation19,102 (A) mAb #9 binding to the outer surface of the hexavalent capsomere. (B) mAb 5B6 binding to the 2 L1 molecules of the adjacent hexavalent capsomeres. (C) A 3D reconstruction structure of mAb H11.B2 binding to HPV11 VLP (the DE loops on L1, ) indicates that the binding sites are located at the center of the capsomere. (D) A 3D reconstruction structure based on cryo-EM data of mAb H16.V5 binding to HPV16 VLP (FG and HI loops on L1, ) shows that H16.V5 only binds to the hexavalent capsomeres but not the pentavalent capsomeres.Citation19 Recently, atomic model of the V5 epitope were built with higher resolution 3D reconstruction of cryo-EM data, demonstrating certain conformational changes induced by V5 binding to its epitope and confirming the preferential binding to the hexavalent capsomeres.Citation151 The different binding models demonstrate different neutralization mechanisms for neutralizing antibodies exerting their effects against the viral infection. (E) The 3D structural model of mAb H11.B2 binding to HPV VLP in which the binding site of H11.B2 was indicated in blue and warmpink for VLP. (F) The 3D structural model of mAb H16.V5 binding to HPV16 VLP, with magenta for H16.V5 binding site on FG loop and red on HI loop, and cyan for VLP.