Figures & data
Figure 1. Induction of antiviral gene expression in PBS-12SF cells by stimulation with different amounts of recombinant chicken IFNα (chIFNa). Total RNA was isolated from PBS-12SF cells after 4 and 8 h induction with 0, 10, 100 and 400U chIFNa. Cells were harvested at 4 and 8 h post induction. RNA was isolated for synthesis of cDNA. Abundance of various mRNAs was assessed using 30ng of cDNA by quantitative real-time RT-PCR (qPCR) analysis with SYBR Green. Genes studied included: IFIH1, Mx1, OAS, IRF3, and PKR. β-actin was used to normalize relative expression, and data was analyzed by the 2ΔΔCt method.Citation20 Mean values and standard deviations of 2 independent experiments are shown. Significance was calculated by Student's t test (*, P ≤ 0.05 and **, P ≤ 0.01).
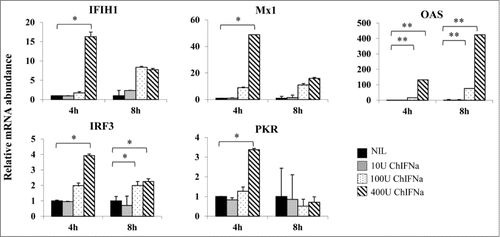
Figure 2. INF response and antiviral gene expression in PBS-12SF cells following infection with 2 influenza A viruses. Cells were infected with the indicated virus at an MOI of 0.1. Cells were harvested at 24 and 48 h post-infection, and RNA was isolated for synthesis of cDNA. Relative mRNA (mRNA) abundance was determined by qPCR using specific primers (see ) for IFNα, IFNβ, OAS, IFIH1, and Mx1 genes. Data was analyzed by the 2ΔΔCt methodCitation20 using β-actin as an endogenous control. Mean values and standard deviations of 3 independent experiments are shown. Significance was calculated by Student's t test (*, P ≤ 0.05 and **, P ≤ 0.01).
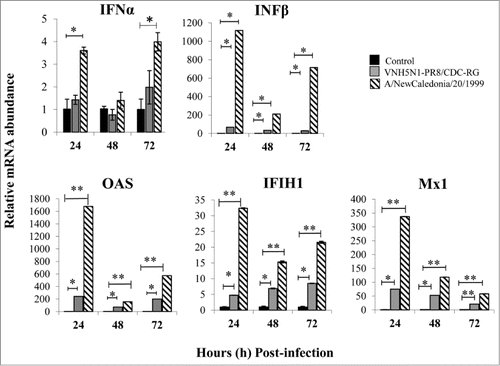
Table 1. Primers used for qPCR and shRNA sequences used for knocking down expression of IFNAR1
Figure 3. Lentiviral-mediated shRNA silencing of IFNAR1 reduces the mRNA abundance of IFN stimulated genes (ISG) in PBS-12SF cells. (A) Characterization of IFNAR1 antibody by peptide-competition assay. PBS-12SF cellular lysates were loaded on a 12% acrylamide gel and transferred to a PDVF membrane for detection by Western blot. Competition was performed by pre-incubating rabbit polyclonal chicken IFNAR1 antibody with increasing amounts (0, 0.1, 0.5, 1 (not shown) or 10 μg) of the immunizing peptide prior to use in Western blotting. A band at approximately 60 kDa was competed out with increasing amounts of peptide (rectangular box). An additional band at approximately 120 kDa was not competed, suggesting this was a non-specific binding. (B) IFNAR1 expression was knocked down in PBS-12SF cells using 3 shRNAs against IFNAR1. Representative Western blot analyses for IFNAR1 in lysates derived from parental PBS-12SF cells and lentiviral-transduced PBS-12SF cells was performed. β-actin was used as a loading control. (C) The mean intensity of the IFNAR1 protein band in (B) expressed as a ratio of the intensity of β-actin. The results shown are the mean of 2 separate experiments. (D) IFNAR1 knockdown reduces the mRNA abundance of OAS, IRF3 and IFIH1 after 4 hours induction with 400U chIFNα. Mean values and standard deviations of 3 independent experiments are shown. Significance was calculated by Student's t test (*, P ≤ 0.05 and **, P ≤ 0.01).
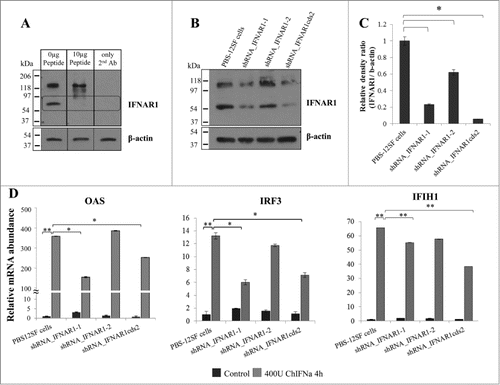
Figure 4. Comparison of the mRNA abundance of hemaglutinin gene in H1N1-infected parental PBS-12SF cells and PBS-12SF cells transduced with IFNAR1-shRNA lentiviruses. Cells were infected at a MOI of 0.1 for 48 h, and supernatants were collected. Viral RNA was purified and qPCR was performed to detect the viral hemaglutinin cDNA. The hemaglutinin gene copy number per 140 µL of culture supernatant is represented in . Mean values and standard deviations of 3 independent experiments are shown. Significance was calculated by Student's t test (*, P ≤ 0.05).
Table 2. Hemagglutination capability was compared in the human influenza virus strain A/NewCaledonia/20/1999 H1N1-infected PBS-12SF and PBS-12SF-IFNAR1-shRNA expressing cells. Cells were infected with influenza at a MOI of 0.1 Supernatants were collected at 48h post-infection, and viral titer measurements were determined
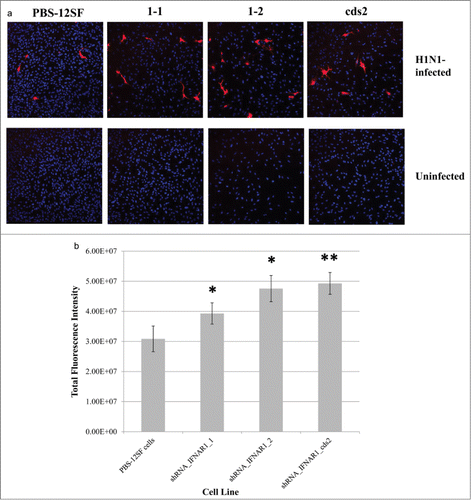
Figure 5. Immunofluorescence of H1N1-infected parental PBS-12SF cells and PBS-12SF cells transduced with 3 separate IFNAR1 shRNA expressing lentiviral vectors. (A) Parental PBS-12SF cells and IFNAR1-shRNA cells were stained with DAPI (blue) and anti-Influenza A H1N1 nucleoprotein (red) as described in Materials and Methods. Images are representative of 10 fields from infected cells and 3 fields from uninfected cells. Imaging was performed at 20× with an Olympus FluoView FV1000 confocal laser scanning microscope. (B) Relative fluorescence in PBS-12SF cells and in PBS-12SF cells transduced with 3 IFNAR1 shRNA expressing lentiviral vectors. Fluorescence values are sums of regions of interests drawn around red-fluorescing cells in 10 fields from each infected slide chamber depicted in (C). An * indicates P < 0.05 and ** indicates P < 0.01.