Figures & data
Figure 1. Construction and representative expression of dbDNA™ PR8 and pDNA PR8 constructs. (A) Process of enzymatic production of dbDNA™. Rolling circle amplification of the double-stranded DNA template results in concatamers that are cleaved and joined by the protelomerase TelN to yield the covalently closed, double-stranded cassette. (B) Schematic of the linear double-stranded dbDNA™ PR8 construct with end terminal single-stranded DNA hairpins. The end product was treated with restriction enzymes and exonuclease to remove plasmid backbone sequences. (C) Schematic of pDNA PR8 construct, PR8.HA.ECRO. ECRO refers to the gene sequence containing an IgE leader sequence [e] and the target sequence is codon [c] and RNA [r] optimized [o]. The sequence of PR8 was cloned into the pVax1 mammalian expression vector. The CMV promoter, HA gene, BGH poly A signal, kanamycin resistance gene, and pUC origin are shown. (D) Representative in vitro expression of the dbDNA™ PR8 and pDNA PR8 constructs. Expression was confirmed using transfected RD cells and a HA-tagged antibody. An empty vector (pVax) was used as a negative control. Results were analyzed with confocal imaging. Expression is indicated by fluorescein isothiocyanate (FITC) staining (green).
![Figure 1. Construction and representative expression of dbDNA™ PR8 and pDNA PR8 constructs. (A) Process of enzymatic production of dbDNA™. Rolling circle amplification of the double-stranded DNA template results in concatamers that are cleaved and joined by the protelomerase TelN to yield the covalently closed, double-stranded cassette. (B) Schematic of the linear double-stranded dbDNA™ PR8 construct with end terminal single-stranded DNA hairpins. The end product was treated with restriction enzymes and exonuclease to remove plasmid backbone sequences. (C) Schematic of pDNA PR8 construct, PR8.HA.ECRO. ECRO refers to the gene sequence containing an IgE leader sequence [e] and the target sequence is codon [c] and RNA [r] optimized [o]. The sequence of PR8 was cloned into the pVax1 mammalian expression vector. The CMV promoter, HA gene, BGH poly A signal, kanamycin resistance gene, and pUC origin are shown. (D) Representative in vitro expression of the dbDNA™ PR8 and pDNA PR8 constructs. Expression was confirmed using transfected RD cells and a HA-tagged antibody. An empty vector (pVax) was used as a negative control. Results were analyzed with confocal imaging. Expression is indicated by fluorescein isothiocyanate (FITC) staining (green).](/cms/asset/7ea7a2b9-a0bb-4588-936e-d90dfac0ed6d/khvi_a_1022008_f0001_c.gif)
Figure 2. Study timeline. BALB/c mice immunized with either dbDNA™ PR8 or pDNA PR8 received the treatments listed at the indicated weeks.
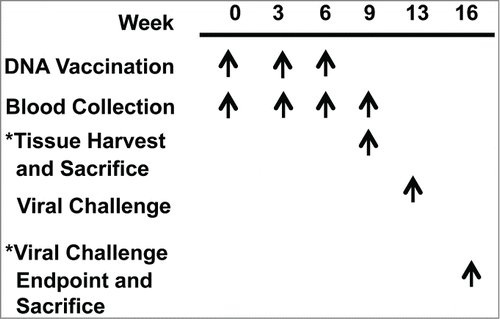
Figure 3. DNA vaccination induces robust antibody responses. Comparison of the antibody responses induced by each vaccination. Absorbances and geometric mean endpoint titers against A/PuertoRico/8/34 are shown for both the dbDNA™ PR8 (A and C) and pDNA PR8 (B and D) constructs. Each vaccination is noted as Dose 1, Dose 2, or Dose 3.
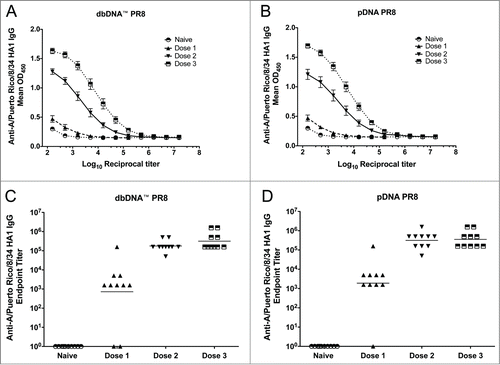
Figure 4. Comparison of host responses to DNA vaccination. (A) Hemagglutination inhibition (HI) antibody titers from the challenge groups of mice (n = 10/group) 3 weeks after the final immunization and prior to viral challenge. (B) The number of IFN-γ secreting splenocytes 3 weeks after the final vaccination. Splenocytes were stimulated with H1HA pooled peptides and IFN-γ secretion was reported as spot forming units (SFU) per million splenocytes. Group mean ± SEM are reported. ***, P < 0.0005 for DNA HI antibody titers compared to naive.
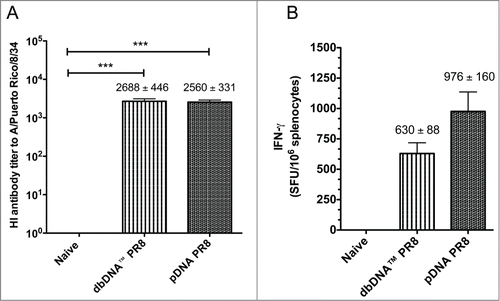
Figure 5. Cytokine frequencies and phenotypic profiles of specific CD4+ and CD8+ T cells following DNA immunizations. Cytokine recall responses to dbDNA™ PR8 and pDNA PR8 were measured 3 weeks after the final immunization by ICS and flow cytometry. Multiparameter flow cytometry was used to determine the percentages of multifunctional CD4+ T cell (A–C) and CD8+ T cell (D–F) cytokines. The graphs show the percentages of each cytokine subpopulation to H1HA pooled consensus peptide stimulation. Background staining from cells stimulated with medium alone has been subtracted. Data represent the mean ± SEM of 5 mice per group. Cytokine responses for animals vaccinated with dbDNA™ PR8 or pDNA PR8 were compared to naïve. ***, P < 0.0005; **, P < 0.005; *, P < 0.05.
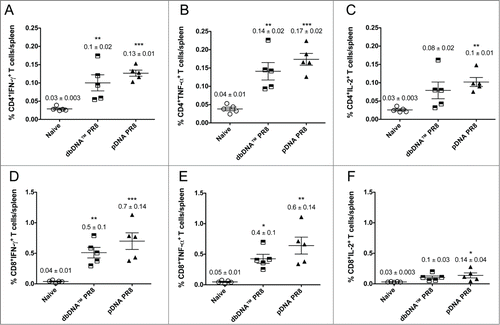
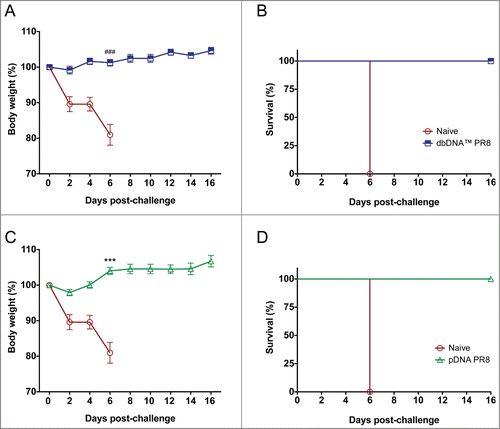
Figure 6. Protection from H1N1 A/Puerto Rico/8/34 virus challenge in DNA immunized mice. Percentage body weight loss following lethal viral challenge with 50 LD50 PR8 Cambridge virus and Kaplan-Meier survival curve showing percentage survival of mice from each group. (A and B) Percentage body weight loss and survival for dbDNA™ PR8 immunized mice. (C and D) Percentage body weight loss and survival for pDNA PR8 immunized mice. Group mean and SEM are reported. ###, P < 0.0005 for dbDNA™ PR8 body weight (%) compared to naïve. ***, P < 0.0005 for pDNA PR8 body weight (%) compared to naïve.