Figures & data
Figure 1. Comparison of GP-specific IgG responses of macaques vaccinated within individual or combination DNA vaccine over time. Groups of 6 cynomolgus macaques were vaccinated 3 times at 4 week intervals with individual optimized MARV (A-B) or EBOV (C-D) DNA vaccines or with a combination of all 4 filovirus vaccines (E-L) as part of 2 separate challenge studies (MARV study, EBOV study). Sera were collected from macaques prior to each vaccination (days 0, 28, 56) and 28 d after the final vaccination (day 84). IgG antibodies were assayed by indirect ELISA using HEK 293T-expressed and purified, soluble antigen representing full length MARV or EBOV GP minus the transmembrane regions (MARV GPΔTM antigen: A, E, I; EBOV GPΔTM antigen: C, G, K) or minus both the transmembrane region and the mucin domain (MARV GPΔMuc antigen: B, F, J; EBOV GPΔMuc antigen: D, H, L). Sera from single agent vaccine groups were tested against the homologous GP antigens only (A-D). To determine the antibody units/ml (Ab U/ml), each serum sample was compared to a standard reference curve of pooled cynomolgus macaque immune sera. The Ab U/ml for each dilution of the test NHP sera were interpolated from this standard curve. Black horizontal bars indicate the mean titers for each group. To evaluate differences in ELISA titers within vaccine groups over time, a repeated measures 2-way ANOVA with Tukey's multiple comparison test was used. Significant differences between study days are indicated (*) and P values for comparisons between days are: 0 vs 56 P < 0.0001; 56 vs 84, P < 0.0001 (A). 0 vs 28 P = 0.0086; 28 vs 56 P = 0.0056; 56 vs 84 P < 0.0001 (B). 0 vs 56 P = 0.0007 (C). 0 vs 56 P = 0.0015; 56 vs 84 P = 0.0048 (D). 0 vs 84 P = 0.0030 (E). 0 vs 56 P = 0.0140 (F). 0 vs 56 P = 0.0123; 56 vs 84 P = 0.0009 (G). 0 vs 56 P = 0.0048; 56 vs 84 P < 0.0001 (H). 0 vs 56 P < 0.0001 (I). 0 vs 56 P = 0.0003 (J). 0 vs 56 P = 0.0484 (K). 0 vs 56 P = 0.0167 (L).
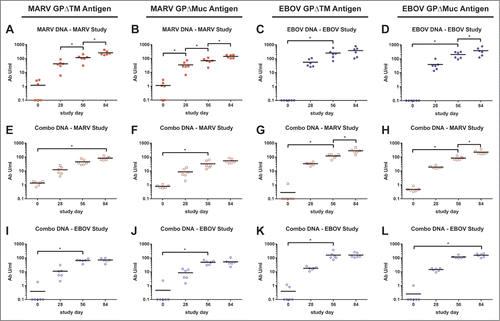
Figure 2. Comparison of GP-specific IgG responses of macaques between vaccine groups over time. These are the same indirect ELISA data shown in , but with individual and combination vaccine groups for within the (A-B) MARV and (C-D) EBOV studies plotted together. Black horizontal bars indicate the mean titers for each group. To evaluate differences in ELISA titers between vaccine groups over time, a repeated measures 2-way ANOVA with Tukey's multiple comparison test was used. Significant differences between vaccine groups are indicated (*) and P values for comparisons between days are: (A) day 56, P = 0.0170; day 84, P < 0.0001. (B) day 56, P = 0.0118; day 84, P < 0.0001. (C) day 84, P = 0.0088. (D) day 84, P = 0.0006.
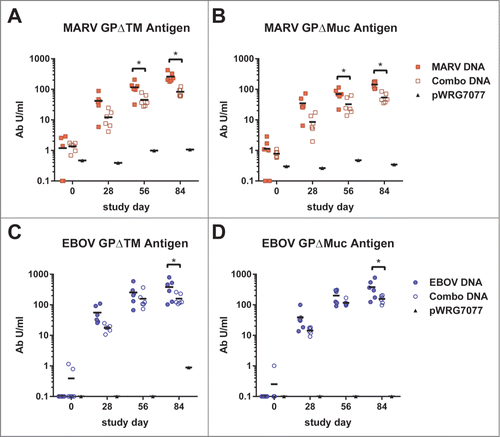
Figure 3. Neutralizing antibody responses elicited by filovirus DNA vaccines in macaques. A dilution series of heat-inactivated sera (day 84) supplemented with human complement was incubated with VSVΔG*rLuc pseudovirions decorated with the glycoproteins of each filovirus and then used to infect Vero cells. At 18–24 hrs post-infection, the cells were lysed, luciferase substrate was added, and the resulting signal was measured using a luminometer. The data were fit to a sigmoidal dose/response curve with variable slope to determine the serum titer required to neutralize infection by 50% relative to untreated controls (PsVNA50), and the limit of quantitation for this assay was 1.3 logs. The log10 PsVNA50 titers are shown for the MARV (A) and EBOV (B) studies. There was no significant difference (ns) in the mean log10 PsVNA50 titers against MARV PsVs between the MARV-GP and combination vaccine groups as determined by an unpaired 2 tailed t-test, P = 0.0702. Similarly, there was no significant difference in the mean log10 PsVNA50 titers against EBOV PsVs between the EBOV-GP and combination vaccine groups as determined by an unpaired 2 tailed t-test, P = 0.3210.
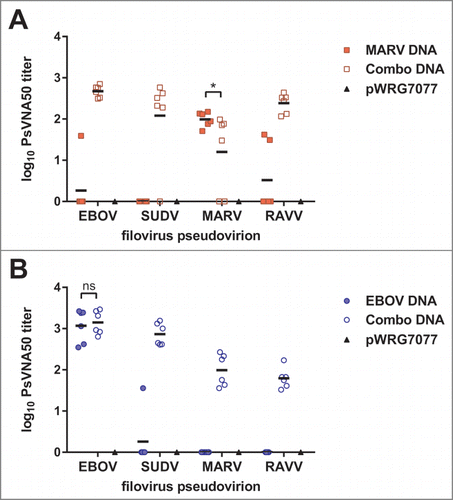
Figure 4. IFN-γ producing cellular responses elicted by filovirus DNA vaccines in macaques. Cryopreserved peripheral blood mononuclear cells collected on day 56 were revived and stimulated with (A) MARV- or (B) EBOV-derived peptides spanning their respective GP proteins for 48 hrs. Peptide-specific memory T cell responses were assayed by IFN-γ ELISpot. Black horizontal bars indicate the means for each group. There was no significant difference (ns) between single and combination vaccine groups in either study unpaired 2 tailed t-test (MARV study, P = 0.8263, EBOV study, P = 0.8171)
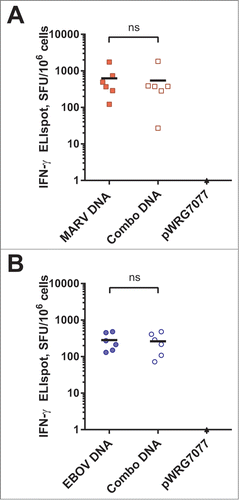
Figure 5. Protective efficacy of filovirus DNA vaccines and clinical observations in MARV-challenged macaques. (A) Kaplan-Meier survival curves were generated for the MARV vaccination study. A Fisher's exact test with multiple comparison adjustment based on permutation was used to evaluate pairwise differences (P < 0.05) relative to the control group (*). Historical controls (†) that received the same MARV challenge dose and route of administration were included for statistical analyses (n = 3). (B-D) After MARV challenge, macaques were scored twice daily for clinical signs typical of hemorrhagic fever disease such as weight loss, changes in body temperature, petechial rash, ischuria, gastrointestinal distress, labored breathing, lack of responsiveness, recumbency, edema, and anorexia. Personnel performing observations were blinded to the treatment groups.
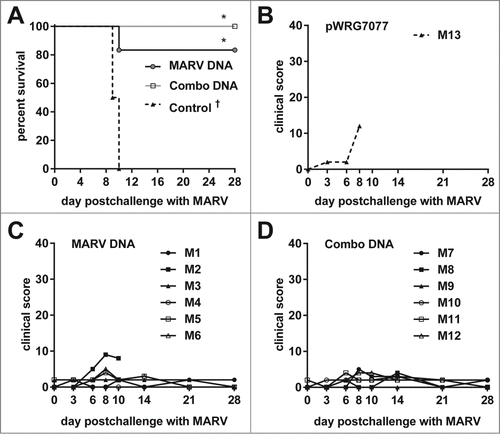
Figure 6. Protective efficacy of filovirus DNA vaccines and clinical observations in EBOV-challenged macaques. (A) Kaplan-Meier survival curves were generated for the EBOV vaccination study. A Fisher's exact test with multiple comparison adjustment based on permutation was used to evaluate pairwise differences (P < 0.05) relative to the control group (*) or to both the control and combination groups (∴). Historical controls (†) that received the same EBOV challenge dose and route of administration were included for statistical analyses (n = 15). (B-D) After EBOV challenge, macaques were scored twice daily for clinical signs typical of hemorrhagic fever disease such as weight loss, changes in body temperature, petechial rash, ischuria, gastrointestinal distress, labored breathing, lack of responsiveness, recumbency, edema, and anorexia. Personnel performing observations were blinded to the treatment.
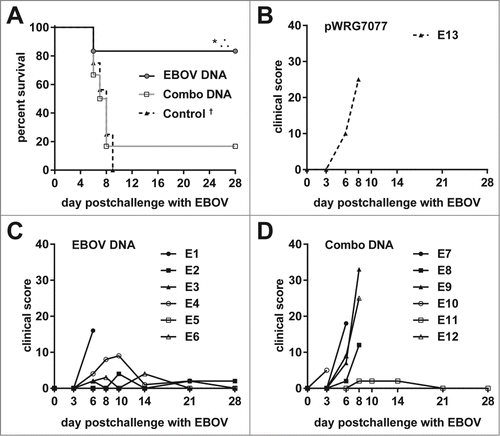
Figure 7. Viremia, AST levels, and platelet counts of MARV-challenged macaques. Whole blood was collected from nonhuman primates at the indicated time points post challenge with MARV. The measured parameters for each animal are shown grouped by vaccine that was administered: (A, D, G) EBOV-GP; (B, E, H) Combination; (C, F, I) pWRG7077. Serum viremia was determined by standard filovirus plaque assays.
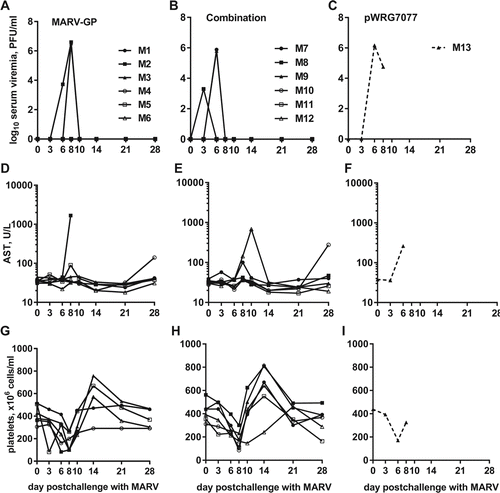
Figure 8. Viremia, AST levels, and platelet counts of EBOV-challenged macaques. Whole blood was collected from nonhuman primates at the indicated time points post challenge with EBOV. The measured parameters for each animal are shown grouped by vaccine that was administered: (A, D, G) EBOV-GP; (B, E, H) Combination; (C, F, I) pWRG7077. Serum viremia was determined by standard filovirus plaque assays.
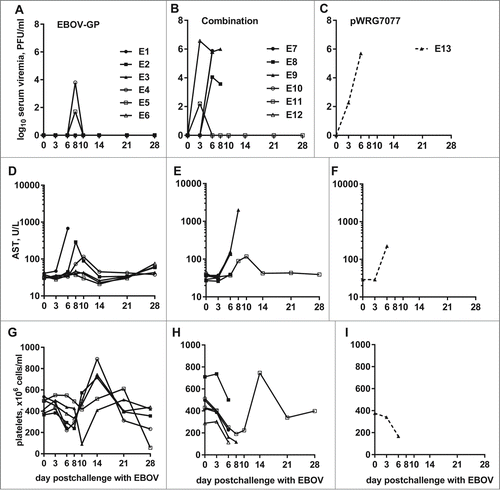
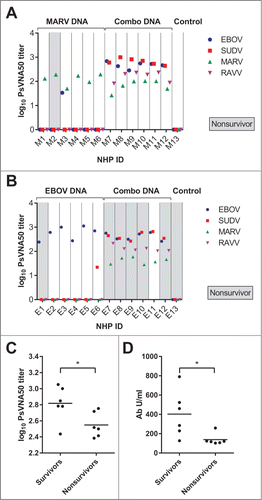
Figure 9. Survival from EBOV challenge correlates with increased antibody responses in cynomolgous macaques. These are the same PsVNA data shown in , but with the data regrouped by individual NHP in the (A) MARV or (B) EBOV study groups. Animals which succumbed to challenge are highlighted in gray (“Nonsurvivor”). (C) EBOV PsVNA data for the EBOV study group were additionally regrouped by survival. (D) These are the same EBOV GPΔTM-specific ELISA data shown in regrouped by survival. Black horizontal bars indicate the mean of each group. There was a significant difference in the mean log10 PsVNA50 titers against EBOV PsVs between the survivor and nonsurvivor groups as determined by an unpaired 2 tailed t-test, p = 0.0331. There was also a significant difference in the mean ELISA titers between these groups as determined by a repeated measures 2-way ANOVA with Sidak's multiple comparison test.