Figures & data
Table 1. Drugs used to treat cells and assay for alterations in the CLIP/HLA-DR ratios.
Table 2. Correlation of cell death and reduction in CLIP/HLA-DR ratios.
Figure 1. Effect of drug panoply on the CLIP/HLA-DR ratio. Y axis = [Mean fluorescence intensity (MFI) of anti-CLIP minus the MFI of the ant-CLIP isotype control] divided by [MFI of anti-HLA-DR minus the MFI of anti-HLA-DR isotype control]. X axis represents the drug panoply, with individual drugs indicated. (A) Raji; (B) Raji repeat, but note: DMSO was not done for this experiment; (C) HLA class II-negative, RJ subclone of Raji. All MFI values represent gating on live cells.
![Figure 1. Effect of drug panoply on the CLIP/HLA-DR ratio. Y axis = [Mean fluorescence intensity (MFI) of anti-CLIP minus the MFI of the ant-CLIP isotype control] divided by [MFI of anti-HLA-DR minus the MFI of anti-HLA-DR isotype control]. X axis represents the drug panoply, with individual drugs indicated. (A) Raji; (B) Raji repeat, but note: DMSO was not done for this experiment; (C) HLA class II-negative, RJ subclone of Raji. All MFI values represent gating on live cells.](/cms/asset/87b99210-ea25-4faf-861a-55e73a424e4b/khvi_a_1089370_f0001_oc.gif)
Figure 2. Assessment of CLIP alone for drug effects. (A) Average of data above; (B) Average of anti-CLIP MFI alone (without subtracting isotype control; without obtaining a ratio with anti-HLA-DR MFI). (C) MFI for anti-CLIP for the T5–1 B-cell line, with the indicated drugs. (D) MFI for anti-CLIP for the HLA class II negative 616 Citation7 subclone of T5–1.
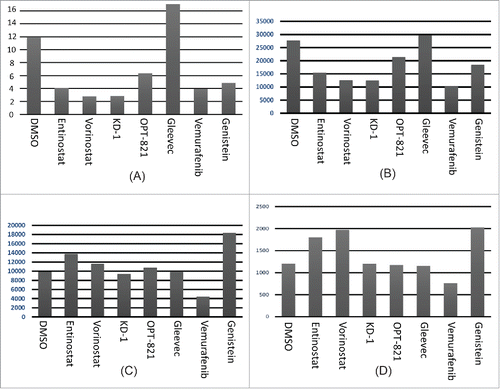
Figure 3. Percent of live cells following drug treatments. Drug treatments as indicated on the X-axis. (A) Data taken from experiment represented by . (B) Data taken from experiment represented by .
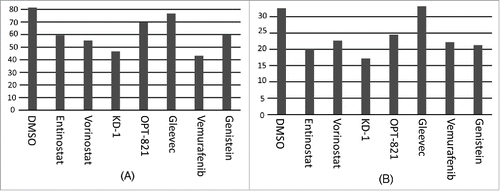
Figure 4. Sensitivity of IEDB, HLA-DR bacterial epitopes to proteases. Proteases indicated along the X-axis; sensitivity along the Y-axis. Bars above zero represent proteases where the average sensitivity of all the IEDB bacterial epitopes is greater than the average sensitivity of all the epitopes for all the proteases. Bars below zero indicate proteases where the average sensitivity of all the IEDB bacterial epitopes is less than the average for the entire set of proteases. For example, most IEDB bacterial epitopes are relatively resistant to matrix metalloprotease-12 (MMP-12), consistent with high level MMP-12 expression in macrophage.Citation2,13 And, most IEDB bacterial epitopes are relatively resistant to Cathepsin L, B and S, consistent with the role of these proteases in antigen processing.Citation3,6,8,11
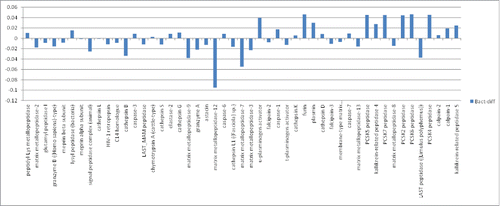
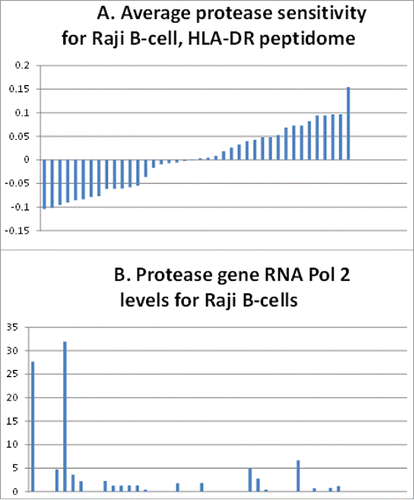
Figure 5. Correlation of Raji B-cell, protease gene, pol 2 levels with the HLA-DR peptidome epitopes' resistance to the proteases. (A) Sensitivity (above the average midline) or resistance (below the average midline) of epitopes of the Raji B-cell HLA-DR peptidome determined in ref.Citation3 (B) Levels of RNA polymerase 2 associated with the protease genes representing the above bars in the histogram of (A).